Cecilia Lu
Cecilia S. Lu, Ph.D.
Research Scientist
Science and Technology Group
Affiliated with Formation and Regulation of Neuronal Connectivity Research Unit
Office: Lab1 C-408 Phone: 098-966-8390; FAX: 098-966-2890 Email: cecilia.lu@oist.jp
Cecilia Lu joined OIST in spring 2012 and has been a Tutor for the OIST Developmental Neurobiology Course. Since completion of Ph.D. in Neuroscience from Brandeis University in 2004, Cecilia has been investigating cellular and molecular mechanisms that regulate neuromuscular synapse development in Drosophila with a focus on microRNAs. Between 2005 and 2011, Cecilia conducted post-doctoral research in the Department of Cell Biology in Harvard Medical School. During her graduate training from 1998 to 2004, Cecilia studied learning and memory from the biochemical perspective of Calcium/Calmodulin-dependent Protein Kinase II (CaMKII) and its regulation by synaptic scaffolding protein CASK. Cecilia became interested in neurobiology, protein kinases, and functional RNAs while she worked as an undergraduate research intern in Academia Sinica in Taiwan and in Yale University School of Medicine. She graduated from National Tsing Hua University in Taiwan in 1997 with a B.S. in Life Science.
MicroRNA Functions in Synapse Development: Epigenetic instructions for how to micro-manage neuronal connectivity and achieve plasticity and stability
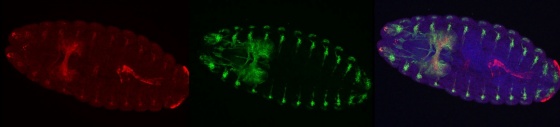 Neuronal and neuromuscular connectivity visualized by fluorescent antibody labeling in Drosophila embryo
Neuronal and neuromuscular connectivity visualized by fluorescent antibody labeling in Drosophila embryo
Accurate and timely formation of neuronal connections is critical for neural functions. As neurons acquire the “abilities” to recognize other partner cells and form specific connections called synapses, the expression level of these “abilities” is not regulated solely by a simple ON/OFF switch. Rather, neuronal connectivity in the nervous system is locally and dynamically tuned to generate many IN-BETWEEN states that drive a great degree of developmental and behavioral plasticity.
From a reductionist point of view, the “abilities” of neurons are derived from genes expressing messenger RNAs (mRNAs) that code for the core protein assembly of neural transmission apparatus, ion channels, and receptors/ligands for cell adhesion molecules. While a fraction of genes also express mRNAs that code for transcription factors as ON/OFF switches, the majority serve unknown regulatory functions as non-protein coding RNAs.
MicroRNA (miRNA) is the best studied class of all naturally occurring small non-coding RNAs because it can bind to mRNA with near-perfect complementarity and interferes with mRNA stability or suppresses translation of mRNA into functional proteins. This epigenetic mechanism by which miRNA tunes the expressivity of genes or “abilities” in neurons without altering the genetic code in DNA sequence has generated an intense interest about miRNA functions in many areas of biology including neurogenesis from stem cells. However, little is known about the specific roles played by miRNA in the course of synapse development beyond neurogenesis.
Research Interest
Cecilia Lu is interested in understanding how miRNA contributes to the expressivity of genes involved in target recognition, synapse formation, and synapse stability. She combines a molecular genetic approach with proteomics and imaging of synapses in the developing Drosophila melanogaster neuromuscular junctions (Lu and Van Vactor, Comp Dev Neurosci. 2013) to deliver miRNA-specific interference and to visualize the global and cell-type specific regulation of gene expression. Through quantitative proteomic analysis and genetic interactions, Cecilia Lu and colleagues recently uncovered a subset of cell surface molecules functioning downstream of a conserved miRNA to establish precise target recognition and synapse formation (Lu et al., Philos Trans R Soc Lond B Biol Sci. 2014). Previously, Cecilia Lu and colleagues had identified miRNAs from a genetic screen based on defective synapse development and utilized transgenic artificial miRNA sponges to dissect spatio-temporal specific miRNA functions (Loya and Lu et al., Nat Methods. 2009).
Publications
- Cecilia S. Lu, Alex Mauss, Bo Zhai, Matthias Landgraf, Steve Gygi and David Van Vactor (2014): MicroRNA-8 Promotes Robust Motor Axon Targeting By Coordinate Regulation of Cell Adhesion Molecules During Synapse Development. Philosophical Transactions of the Royal Society B – Biological Sciences, 369 (1652):20130517, August 18, 2014.
- Cecilia S. Lu and David Van Vactor (2013): Genetic analysis of synaptogenesis in: Comprehensive Developmental Neuroscience: Cellular Migration and Formation of Neuronal Connections, edited by John Rubenstein and Pasko Rakic, volume 2, pages 537-577. Academic Press, Elsevier, Inc. ISBN-13: 978-0123972668, June 11, 2013.
- Laura Anne Lowery, Haeryun Lee*, Cecilia S. Lu*, Rebecca Murphy, Robert A. Obar, Jacqueline Rho, Bo Zhai, Steve Gygi, Yougen Zhan and David Van Vactor (2010): Parallel Genetic and Proteomic Screens Identify Msps as a CLASP-Abl Pathway Interactor in Drosophila. Genetics, August 2010, 185(4):1311-25. doi: 10.1534/genetics.110.115626. Epub 2010 May 24. * Co-second authorship.
- Carlos M. Loya*, Cecilia S. Lu*, David Van Vactor and Tudor A. Fulga (2009): Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nature Methods, December 2009, 6(12):897-903. doi: 10.1038/nmeth.1402. Epub 2009 Nov 15. * Co-first authorship. Comment in: Nature Methods, December 2009, 6(12):873-4. doi: 10.1038/nmeth1209-873.
- Cecilia S. Lu and David Van Vactor (2007): Synapse specificity: Wnts keep motor axons on target. Current Biology, Oct 23, 2007, 17(20):R895-8. doi: 10.1016/j.cub.2007.08.029
- Cecilia S. Lu, James J. L. Hodge, Jennifer Mehren, Xiu Xia Sun and Leslie C. Griffith (2003): Regulation of the Ca2+/CaM-Responsive Pool of CaMKII by Scaffold-Dependent Autophosphorylation. Neuron, December 2003, 40(6):1185-97. doi: 10.1016/S0896-6273(03)00786-4.
- Leslie C. Griffith, Cecilia S. Lu and Xiu Xia Sun (2003): CaMKII, an enzyme on the move: regulation of temporospatial localization. Molecular Interventions, October, 2003, 3(7):386-403.




