FY2016 Annual Report
Plant Epigenetics Unit
Assistant Professor Hidetoshi Saze
Abstract
Epigenetic Regulation of Genes and Transposable Elements in Plants
We are trying to understand epigenetic mechanisms for regulation of gene and transposon activities in the genomes of model plant systems such as Arabidopsis thaliana and rice, using genetic, biochemical and genomic approaches. In addition, we are working on a research project called "functional Okinawan rice project” supported by Okinawa prefecture, in which we are producing a functional rice strain for Okinawa using molecular breeding techniques.
1. Staff
- Hidetoshi Saze, Assistant Professor
- Yuji Miyazaki, Researcher
- Kenji Osabe, Researcher
- Matin Miryeganeh, Researcher
- Yoshiko Harukawa, Technical Staff
- Saori Miura, Technical Staff
- Yoko Fujitomi, Research Administrator
- Nino Espinas, Student
- Tomohito Wauke, Student
- Ruth Thompson, Student
2. Collaborations
3. Activities and Findings
Epigenetic Regulation of Intronic Transgenes
Genome defense mechanism in plants epigenetically inactivate repetitive sequences and exogenous transgenes. An epigenetic inactivation mechanism of intronic T-DNA sequence, termed T-DNA suppression, has been observed in Arabidopsis thaliana. However, the molecular basis of establishment and maintenance of T-DNA suppression is poorly understood.
We have demonstrated that in Arabidopsis, T-DNA suppression by introgression of additional T-DNA sequences is provoked by RNA-directed DNA methylation pathway (Osabe et al., 2017). Suppression of intronic T-DNA is associated with massive DNA methylation throughout the T-DNA insertion sites. Stable maintenance of T-DNA suppression was observed for more than 5 generations, that requires IBM2 and other epigenetic modifiers such maintenance methylase MET1 and chromatin remodeler DDM1. This mechanism parallels the masking of intronic TEs IBM2, and suggesting the presence of a genome surveillance mechanism that masks repetitive DNAs intruding into transcription units.
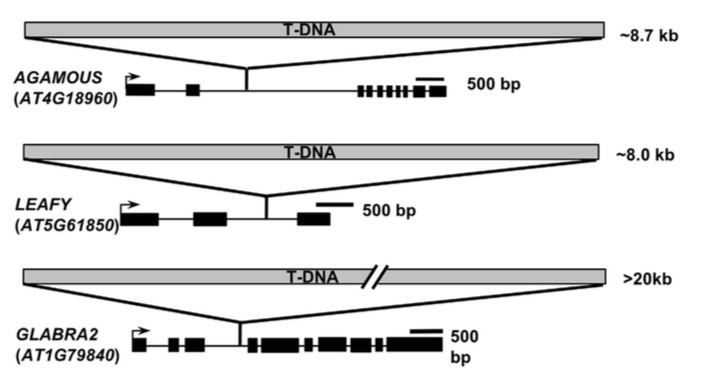

Figure 1: Suppression of T-DNA inserted in AGAMOUS (AG) and LEAFY (LFY) genes. The suppressed ag* and lfy* plants show wild-type flower phenotype even though the presence of >8 kb T-DNA insertion in introns. These suppressions are restored in ibm2 background.
4. Publications
4.1 Journals
-
Osabe K, Harukawa Y, Miura S, Saze H.† (2017). Epigenetic Regulation of Intronic Transgenes in Arabidopsis. Sci Rep. Mar 24;7:45166. doi: 10.1038/srep45166.
4.2 Books and other one-time publications
- (Review) Espinas NA, Saze H, Saijo Y. (2016). Epigenetic Control of Defense Signaling and Priming in Plants. Front Plant Sci. 2016 Aug 11;7:1201. doi: 10.3389/fpls.2016.01201.
4.3 Oral and Poster Presentations
- (Invited talk) Saze H. Epigenetic regulation of intragenic repeats in the plant genomes. The 88th annual meeting of the genetics society of Japan. Nihon University, Mishima, Japan. Sep 7, 2016
- (Invited lecture to graduate students) Saze H. Epigenetic Regulation of Genes and Transposable Elements in Plant Genomes. Sokendai, Hayama, Kanagawa, Japan. February 7, 2017.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
Nothing to report
7. Other
Nothing to report.



