【Seminar】"Enzymatic mechanism of RecA-recombinase in homologous recombination: Role of RecA in homology recognition and the functions of RecA-DNA filaments" by Prof. Takehiko Shibata
Date
Location
Description
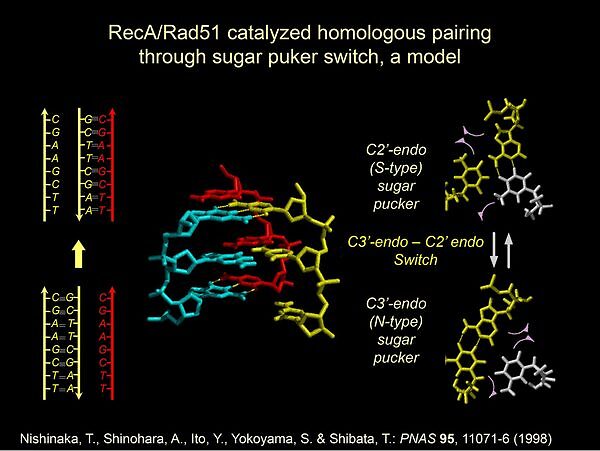
DNA double-strand breaks (DSBs) are often introduced in genomic DNA by radiation, chemicals and even by stress during DNA replication and by oxidative respiration byproducts. DSBs are repaired precisely by homologous recombination, or quickly but less precisely by non-homologous end-joining (NHEJ). Homologous recombination is, thus, essential to maintain genome integrity to prevent diseases and cell death. On the other hand, during meiosis in sexual reproduction, homologous recombination generates genomic diversity, which stimulates the adaptation of each species to environmental changes. Homologous recombination of genomic DNA in virtually all organisms depends on ATP-dependent RecA family recombinases. The known exceptions are homologous recombination by some bacterial viruses and in yeast mitochondria: these types of homologous recombination use ATP-independent recombinases. The RecA-family recombinases include RecA in bacteria, RadA in archaea, Rad51 in eukaryotes from yeasts to human and higher plants, and meiosis-specific Dmc1. In homologous recombination, RecA-family recombinases catalyze ATP-dependent homologous pairing, which is the homologous joint-formation between single-stranded DNA (ssDNA) and an internal complementary sequence of homologous double-stranded DNA (dsDNA). Classic electron microscopy, high resolution cryo-EM and X-ray crystallography revealed a common right-handed spiral filaments of RecA/Rad51 formed around ssDNA or dsDNA. It has long been recognized that the filamentous structure of RecA/Rad51/Dmc1 formed with ssDNA is the instrument to catalyze homologous pairing with dsDNA. Homologous pairing by RecA and Rad51 has been studied, since its discovery in 1979 (Shibata et al. PNAS 1979; McEntee et al. PNAS 1979). Recent high resolution structural studies and single molecule analysis clarified details of the reaction steps of the homologous pairing. However, a critical question in homologous recombination has not been answered; that is, how homologous sequence between ssDNA and dsDNA is recognized to form stable homologous joint with almost perfect sequence match. In this seminar, I will present our findings that in the absence of filament-formation, non polymerizing RecA dimer recognizes and pairs small homologous sequences between ssDNA and dsDNA in an ATP-dependent but hydrolysis-independent fashion. This finding opened an approach to study the homologous pairing intermediates using high resolution structural analysis. On the other hand, an ATP-hydrolysis dependent RecA-function is suggested to plays a role in the accuracy of homologous recombination by exclusion of homologous joints containing mismatch base-pairs.
Attachments
Subscribe to the OIST Calendar: Right-click to download, then open in your calendar application.



