FY2020 Annual Report
Sensory and Behavioural Neuroscience Unit
Assistant Professor Izumi Fukunaga

Abstract
Despite the pandemic, thanks to OIST's PCR team and leadership and some changes enabled by technology, we have been able to work safely. Scientifically, our hard work from the previous years are starting to bear fruit: the molecular study of the olfactory bulb output neurons - a collaboration between OIST, Duke University and Tsukuba University, and a study that started in 2017 - is now under revision. Coming year will, hopefully, be even more exciting with more studies maturing and revealing insights about the brain.
1. Staff
- Dr. Izumi Fukunaga, Principal Investigator
- Dr. Sander Lindeman, Researcher
- Dr. Janine Reinert, Researcher
- Dr. Adam Mago, Researcher (from June 2021)
- Ms. Aliya Adefuin, Graduate Student
- Ms. Xiaochen Fu, Graduate Student
- Ms. Yu-Pei Huang, Technical Staff
- Ms. Sayori Gordon, Administrative Assistant
- Ms. Sourjya Nath, rotation student
- Ms. Rahel Ruppli, rotation student
2. Collaborations
- Dr Hiroaki Matsunami (Duke University)
- Dr Hiroto Sekiguchi (Toyohashi University of Technology)
3. Activities and Findings
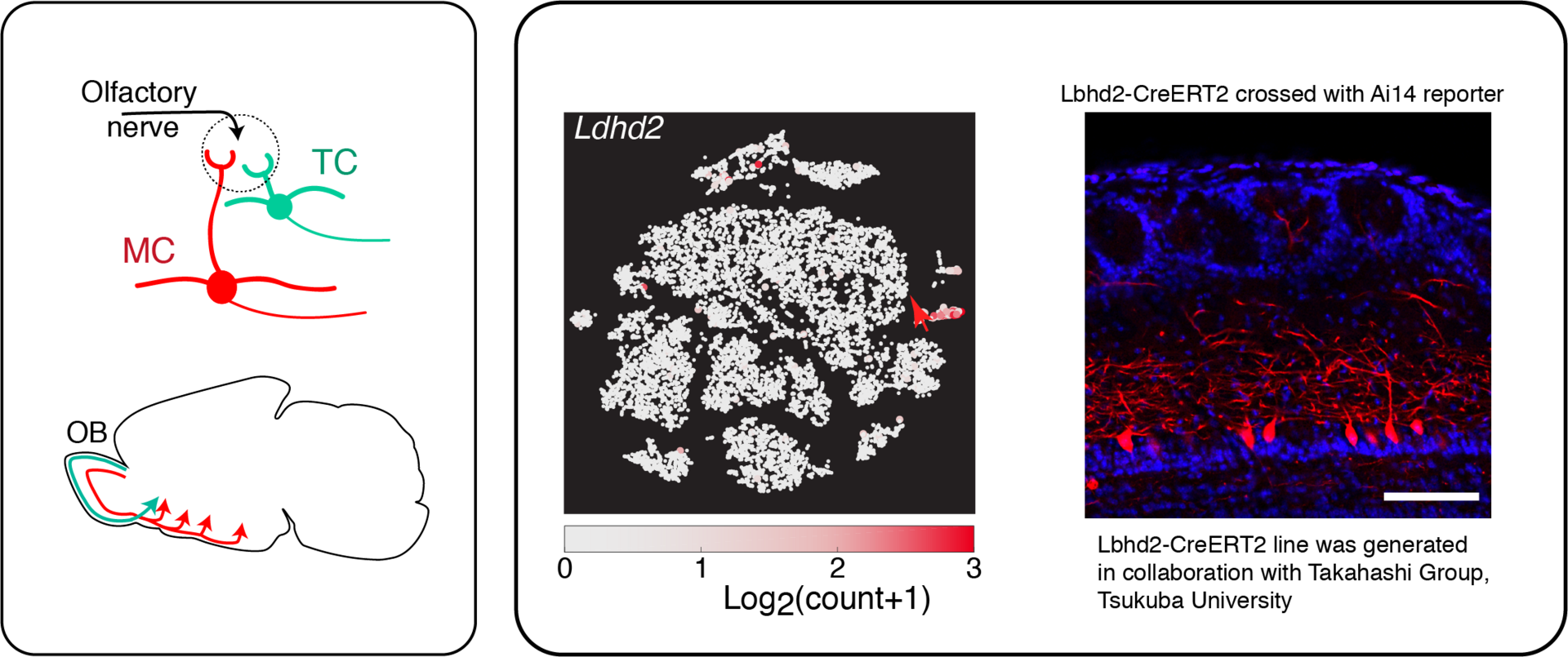
3.1 Molecular study of the olfactory bulb output neurons
A key function of sensory systems in the brain is to extract distinct features from the environment, which often co-exist and implement parallel information processing. Mechanistically, these correspond to unique cell types.
In olfaction, processing of information in the brain starts in the olfactory bulb. Here, the information processing splits into two major pathways, into those carried by mitral cells, and by tufted cells. The former, and larger of the two cell types is of particular interest, due to their wide projection patterns and the long temporal window that is thought to allow mitral cells to carry more complex information. Yet, there has been a lack of selective marker genes or genetic tools to precisely analyse mitral cells and tufted cells. This is the very void that is tackled in the current study. We describe an analysis of publicly available single-cell RNA-seq data in search of unique molecular markers for a key cell type for olfaction, namely the mitral cells, followed by a CRISPR/Cas9-mediated generation of a Cre-driver line using the result, in order that cell-type specific analyses of olfactory processing become possible.
4. Publications
4.1 Journals
Ackels, T., Jordan, R., Schaefer, A.T., and Fukunaga, I. (2020). Respiration-Locking of Olfactory Receptor and Projection Neurons in the Mouse Olfactory Bulb and Its Modulation by Brain State. Frontiers in Cellular Neuroscience 14, 220
Koldaeva, A., Zhang, C., Huang, Y.-P., Reinert, J., Mizuno, S., Sugiyama, F., Takahashi, S., Soliman, T., Matsunami, H., and Fukunaga, I. (2020). Transcriptome analysis for the development of cell-type specific labeling to study olfactory circuits. bioRxiv, 2020.2011.2030.403865; Under revision at a peer-reviewed journal
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Japan Neuroscience online meeting (co-chaired a symposium)
- JSAP (Japan Society for Applied Physics) meeting 2020
- Online ISOT meeting
- OIST-University of Tokyo exchange
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report



