FY2011 Annual Report
Microbiology and Biochemistry of Secondary Metabolites Unit
Holger Jenke-Kodama
Abstract
Our research unit was renamed as “Microbiology and Biochemistry of Secondary Metabolites Unit” in October 2011. The unit was established in May 2009, starting under its original name “Evolutionary Systems Biology Unit”. Though we are still interested in evolutionary aspects of secondary metabolism and systems biology-inspired approaches, we believe that the new name better describes our activities, which, owing to the fact that we have been able to establish several collaborative research projects, have developed on a broader scale than implied by the former name.
The unit is working on both prokaryotic and eukaryotic microorganisms. Research topics include bacterial model organisms, toxin producing dinoflagellates and microbial communities that are associated to marine invertebrates. Moreover, as a collaboration partner within a CREST-sponsored project, we are studying a green microalgal species in order to improve its biotechnological potential as a biofuel source.
In the course of fiscal year 2011 (April 2011 to March 2012), we continued our work on a cyanobacterial model strain and diverse projects that have a focus on marine topics and biotechnological applications. We started to analyse toxin production and gene expression with cultured dinoflagellates and also initiated a study that aims to find correlations between palytoxin content and microbial communities in a marine invertebrate.
Happily, we got the internal funding to raise the number of group members. In addition to the two researchers who joined the unit in 2010 we were able to hire two technicians, Mr. David Richter and Mr. Yosuke Taira, and thus could increase our workforce considerably. This helped enormously to advance in the sampling and labour-intensive projects such as the Ishigaki/palytoxin project and dinoflagellate cultivation. Likewise, our unit profited very much from the newly established mass spectrometry core facility, which is part of the Common Resources Section and became fully operational in April 2011.
We also joined an Okinawa-based project on the effects of an artificial causeway on coastal biodiversity. This project was initiated by Dr. James D. Reimer from the Rising Star Programme of the University of the Ryukyus, the main university in Okinawa, and we were successful in getting external funding for the research work from two private foundations.
In the period from April 2011 to March 2012, we published two primary research papers in international peer-reviewed journals.
1. Staff
- Dr. Holger Jenke-Kodama, Assistant Professor
- Dr. Maiko Tamura, Researcher
- Dr. Kugako Sugimoto, Researcher
- Mr. David Richter, MSc, Technician
- Mr. Yosuke Taira, MSc, Technician
- Ms. Ryoko Uchida, Research Administrator
2. Collaborations
- Theme: Botryococcene biosynthesis in the green microalga Botryococcus braunii
- Type of collaboration: Joint research
- Researchers: Dr. Shigeru Okada, Associate Professor, Graduate School of Agricultural and Life Sciences, The University of Tokyo
- Theme: Microbial communities in Palythoa tuberculosa and palytoxin production
- Type of collaboration: Joint research
- Researchers: Dr. James D. Reimer, Associate Professor, Rising Star Programme, University of the Ryukyus
- Theme: Effects of the Kaichū-Dōro causeway on biodiversity
- Type of collaboration: Joint research
- Researchers: Dr. James D. Reimer, Associate Professor, Rising Star Programme, University of the Ryukyus
3. Activities and Findings
3.1 Toxin biosynthesis in marine dinoflagellates
Dinoflagellates are eukaryotic microalgae that are assumed to produce a variety of marine toxins. The name derives from the Greek word ὁ δῖνος meaning “whirling” or “rotation” and the Latin word flagellum (plural form: flagella) meaning “whip” and refers to a structural characteristic of dinoflagellate cells. About 20 years ago, researchers estimated that of the approximately 1000 extant dinoflagellate species about 30 produced toxins that could be dangerous for humans [1]. However, at that time it was and these days it is still unclear whether the dinoflagellates are the real producers of those compounds or not. The main issue with clarifying this matter is the purity of a culture. Preferably, one would like to work with axenic (Greek prefix ἀ- “un-”/”not” and ξένος “foreign”) cultures, i.e. cultures that contain no "foreign" cells. However, dinoflagellate cultures available from collections are usually not axenic but "contaminated" by other organisms. Therefore, the biosynthetic origin of compounds detected in the culture might be one of the “contaminating” organisms.
We are working with a strain of the dinoflagellate species Ostreopsis ovata (Greek ὂστρεον “oyster- or bivalve-shell”, Greek ὂψις “appearance” and Latin ovatus “egg-shaped”, “oval”) that we received from the former research group of Professor T. Yasumoto in March 2011 after Dr. Tamura had got in contact with this group. The strain was sampled in Okinawa prefecture. Due to the generally very small growth rate of benthic dinoflagellates, it took several months to generate amounts of biomass sufficient for starting experiments. We could confirm by means of liquid chromatography coupled to mass spectrometry (LC-MS) that the strain produces a range of ovatoxin variants. Ovatoxin is structurally closely related to palytoxin (see section 3.2) and supposed to be produced by a polyketide synthase. In the long term, our aim is to elucidate the pathway of palytoxin biosynthesis. For that purpose, one needs to identify the enzymes that are involved in the process as well as the genes they are encoded by. As a first step of this endeavor, we purified total RNA from the cultured cells in order to identify all genes expressed in the toxin-producing dinoflagellate cells. From the total RNA the poly-A-tailed messenger-RNA was isolated, reversely transcribed into DNA and then sequenced using the Illumina “Next generation sequencing” (NGS) platform. We could get sequence data of high quality, which are currently being analysed.
An important issue is that the Ostreopsis ovata culture we are working with is not axenic. Therefore, we cannot exclude the possibility that another organism is the actual producer of ovatoxin. We identified a bacterium as the only organismal contaminant (see Fig. 1) and tried to obtain an axenic dinoflagellate culture by various treatments using antibiotics as well as the oxidizing disinfectant hypochlorous acid. However, these treatments have not been successful yet. We are now trying to identify the bacterium and cultivate it separately.
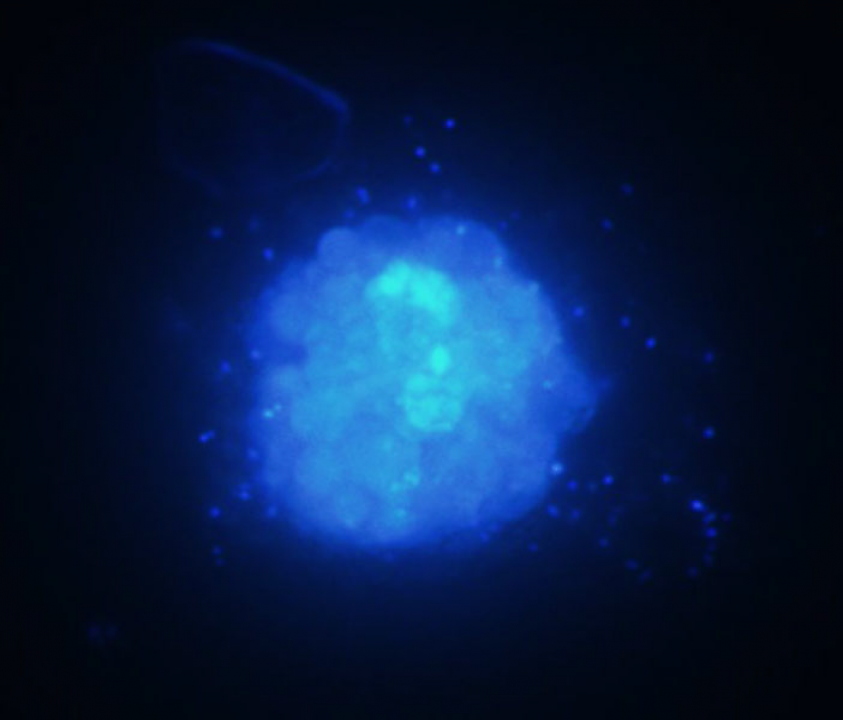
Figure 1. Photomicrograph of the Ostreopsis ovata culture after staining with the DNA-binding fluorescent stain DAPI. This stain can penetrate living cells. After excitation with UV light, DAPI emitts blue light. The photomicrograph shows one O. ovata cell. In the centre, a large amount of densely packed dinoflagellate DNA is visible. The small blue dots are bacteria, which are located in the medium and on the surface of the dinoflagellate cell as well. (Magnification: 600x)
3.2 Microbial communities inside Palythoa tuberculosa and the production of palytoxin
(Collaboration with Dr. James D. Reimer, Associate Professor, University of the Ryukyus)
Palythoa are zoanthids belonging to the subclass Hexacorallia within the class Anthozoa. The name palythoa seems to be derived from Greek πάλυνον “sprinkled” and ὁ θώ as an apocope of ὁ θώραξ “body”, “trunk”. The main palythoa species found in Okinawa is Palythoa tuberculosa [2] (see Fig. 2). They have a simple body plan (see Fig. 2) and grow in colonies in both deep and shallow coral reef ecosystems. Already 40 years ago, a palythoa species was described as the source of palytoxin [3], one of the strongest non-peptide toxins found in nature (see Fig. 3). However, the organism actually producing the toxin has not been identified yet. Bacteria [4] as well as dinoflagellates [5] are possible culprits.


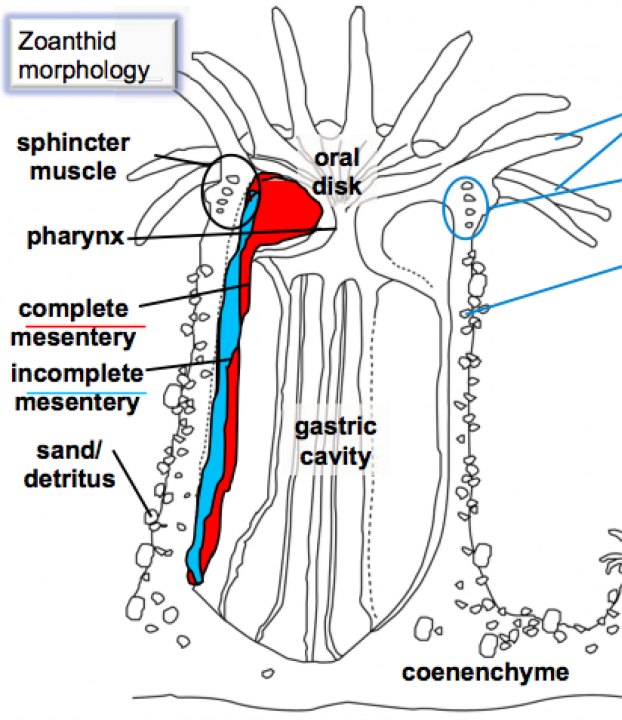
Figure 2. Left: Palythoa tuberculosa colonies. Centre: Detail of an animal. Right: Body plan of a palythoa polyp. (Pictures and drawing kindly provided by Dr. J. D. Reimer)
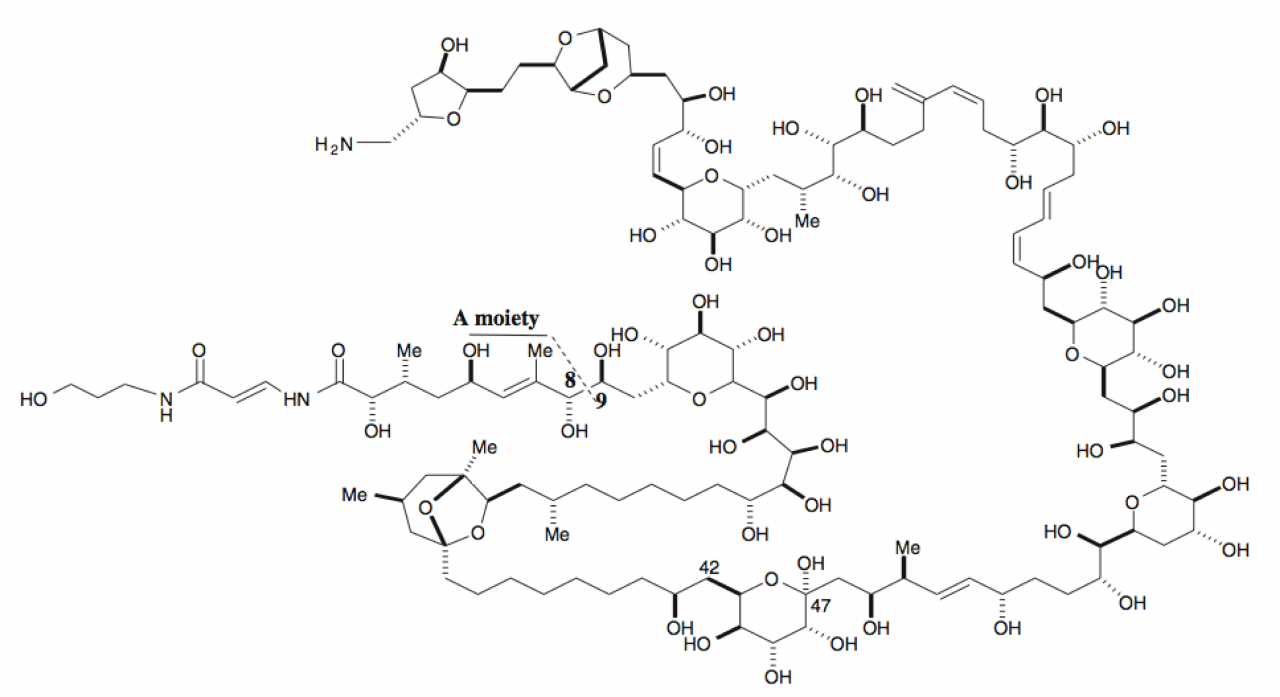
Figure 3. Structural formula of palytoxin.
Starting from April 2011, we have been studying toxin distribution patterns in colonies of Palythoa tuberculosa, which we sampled at various locations in Kabira Bay (see Fig. 4) in Ishigaki island (Okinawa prefecture).

Figure 4. Kabira Bay, Ishigaki, seen from south-east.
Our strategy is to (1) measure the palytoxin content in palythoa colonies by LC-MS, (2) analyse the microbial communities (both prokaryotic and eukaryotic) inside the gastric cavity by means of “next generation” amplicon sequencing, and (3) isolate and cultivate potential producer organisms directly from palytoxin-containing polyps (see the scheme in Fig. 5).
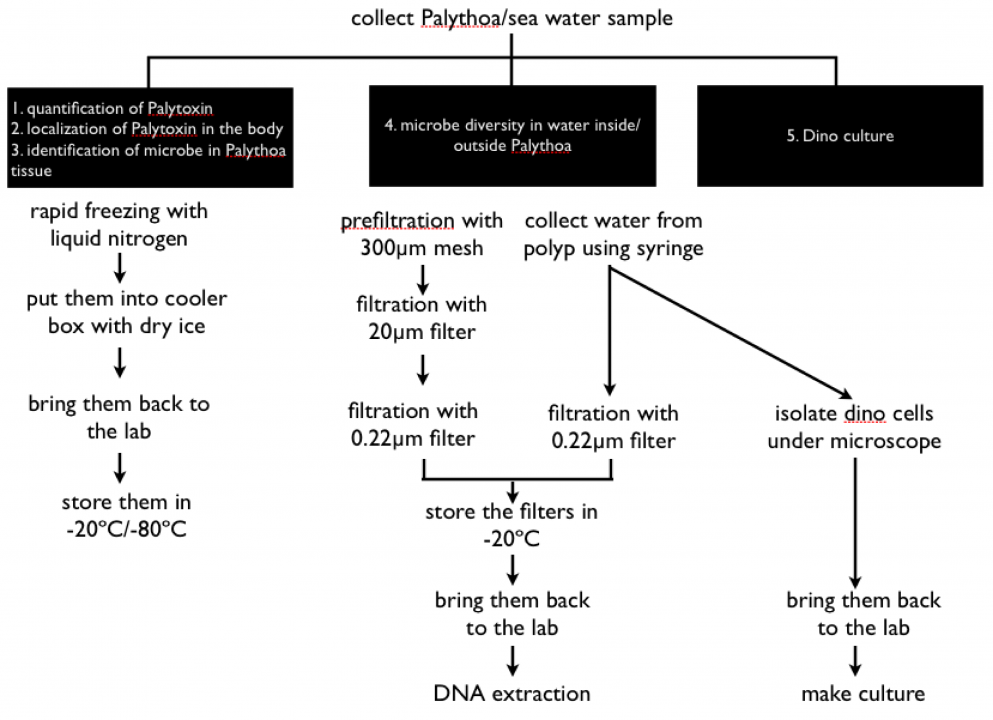
Figure 5. Sample processing scheme for palythoa and seawater samples.
In FY 2011, we sampled together with Dr. James D. Reimer (University of the Ryukyus) palythoa colonies in Ishigaki at three times, namely in April 2011, October 2011 and January 2012. At each point in time, samples were taken at ten different locations and processed for the chemical as well as the sequencing analyses. For each sampling event, palytoxin could be detected by LC-MS in at least one of the samples (see Fig. 6 for an example of a palytoxin-postive mass spectrum) but not in all samples. We found variations in the palytoxin contents hinting at a dependence on the seawater temperature. Furthermore, preliminary analysis of the microbial community data showed that the palytoxin content of colonies might be correlated to the presence of certain bacteria. This is now under investigation. In order to obtain a complete time course, which will also include summer samples, we will be continuing the sampling activities in FY 2012.
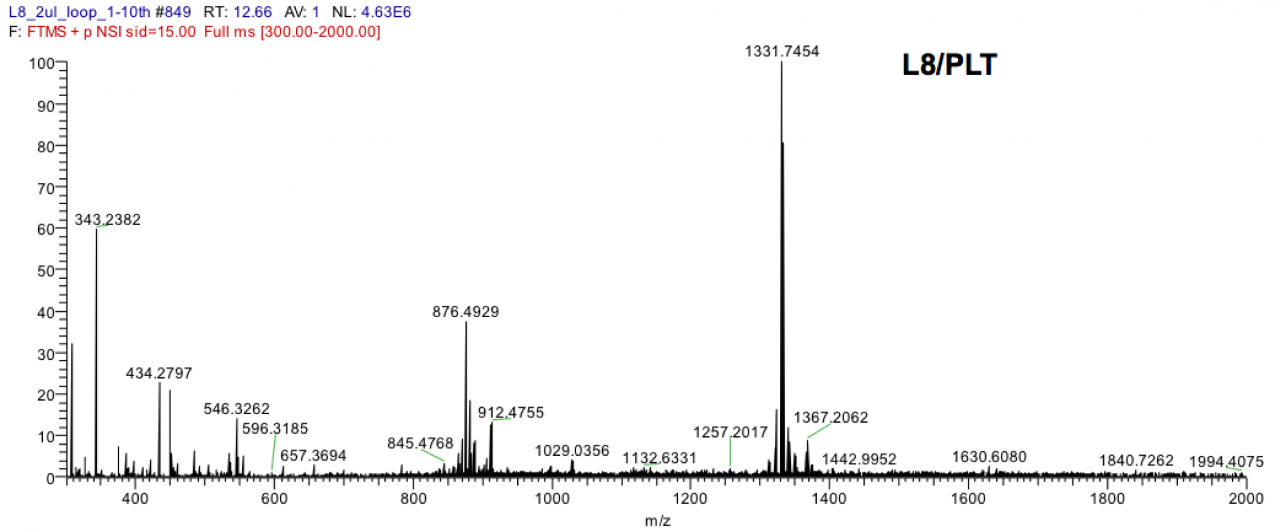
Figure 6. Mass spectrum of a palytoxin-containing Palythoa tuberculosa extract. The peak at a mass to charge ratio of m/z=1331.7454 reveals that palytoxin is present in the sample. (LC-MS measurement performed by Dr. M. C. Roy, Biology Resources Section, OIST)
3.3 Botryococcene biosynthesis in the green microalga Botryococcus braunii
(Collaboration with Dr. Shigeru Okada, Associate Professor, The University of Tokyo)
Botryococcus braunii is a green microalga and belongs taxonomically to the class Trebouxiophyceae within the phylum Chlorophyta [6]. Its name comes from the Greek word ὁ βότρυς, which means “grape” and the Neolatin term coccus for something with a spherical shape. B. braunii is actually a unicellular alga with the individual cells showing a spherical shape, but it grows in colonies, which look like a bunch of grapes. The individual cells are held together by extracellular matrix that contains liquid hydrocarbons in large amounts [7]. The so-called B race produces botryococcenes as the major component [8] (see Fig. 7). Botryococcenes are considered to be a promising source of renewable energy, because their chemical structure is suitable for conventional combustion engines [9].
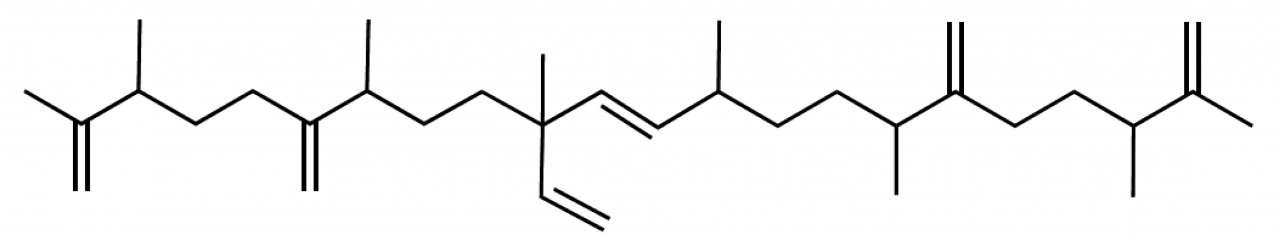
Figure 7. Structural formula of an example botryococcene.
Botryococcenes are triterpenoid hydrocarbons and synthesized from the isoprene units isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). B. braunii produces these isoprene units using a biochemical route that is called MEP pathway (see Fig. 8). This name refers to the compound methylerythritol 4-phosphate (abbreviated as MEP), which is unique to that pathway. To better understand the metabolic flow during the biosynthesis of botryococcenes, it is necessary to characterise the components of the biochemical pathway. The first enzyme of the MEP pathway is 1-deoxy-D-xylulose 5-phosphate synthase (DXS, EC number 2.2.1.7). According to the available genome and experimental data, unicellular microalgae as B. braunii seemed to have only one DXS gene [10], in contrast to land plants, which possess two or more genes encoding the enzyme DXS [11].
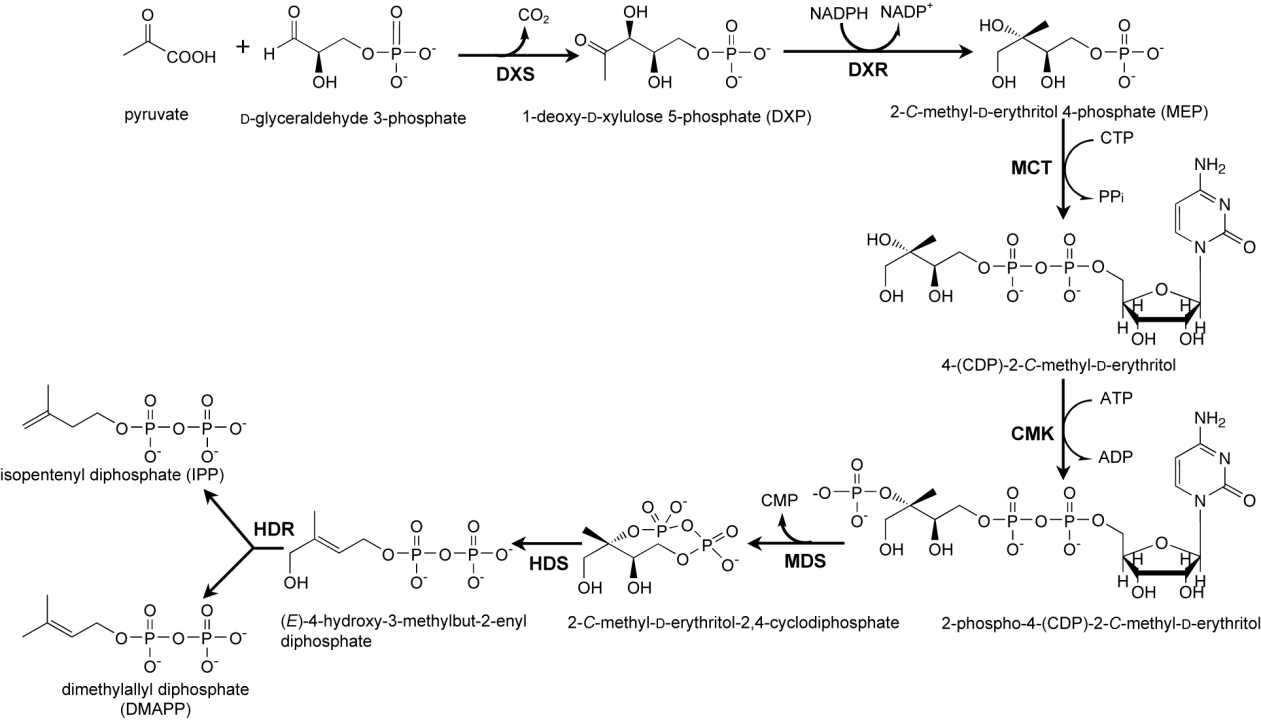
Figure 8. The MEP pathway for the biosynthesis of isoprene units. The enzymes are abbreviated as follows: DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase.
We speculated that, due to the large amounts of botryococcene produced (up to 30% of the algal dry weight), the MEP pathway of B. braunii could be quite unique in order to provide sufficient amounts of isoprene units. Screening of a cDNA library revealed three distinct sequences with high similarity to known microalgal DXS genes. They were named BbDXS-I, BbDXS-II and BbDXS-III. The proteins encoded had calculated molecular masses in the range of 80 kDa. An amino acid alignment with DXS sequences from a land plant (Medicago truncatula, Barrel Clover), another green microalga (Chlamydomonas reinhardtii) and a standard model bacterium (Escherichia coli) showed clear homology and nearly 200 amino acid residues being strictly conserved among all proteins (see Fig. 9). We also reconstructed a phylogenetic tree covering DXS protein sequences from land plants, other green algae, red algae and all major groups of bacteria (see Fig. 10). From this phylogeny, it became clear that the B. braunii proteins did not fit into the three groups described for land plants [12], but rather can be classified as paralogues, which resulted from a gene duplication event.

Figure 9. Amino acid sequence alignment of DXSs. Strictly conserved amino acid residues are shown by shaded backgrounds.

Figure 10. Phylogeny of DXS amino acid sequences, inferred by Bayesian estimation. The node labelled with A represents the last common ancestor of DXS from alpha-proteobacteria and green plants and other photosynthetic eukaryotes. The node labelled with S represents the last common ancestor of DXS proteins from green algae and land plants.
To confirm that the sequences identified really encode functional DXS enzymes, we expressed the proteins heterlogously in the bacterium Escherichia coli, purified them (see Fig. 11) and demonstrated by means of thin-layer chromatography that all three proteins could convert the substrates pyruvate and D-glyceraldehyde 3-phosphate (D-GAP) into the reaction product.
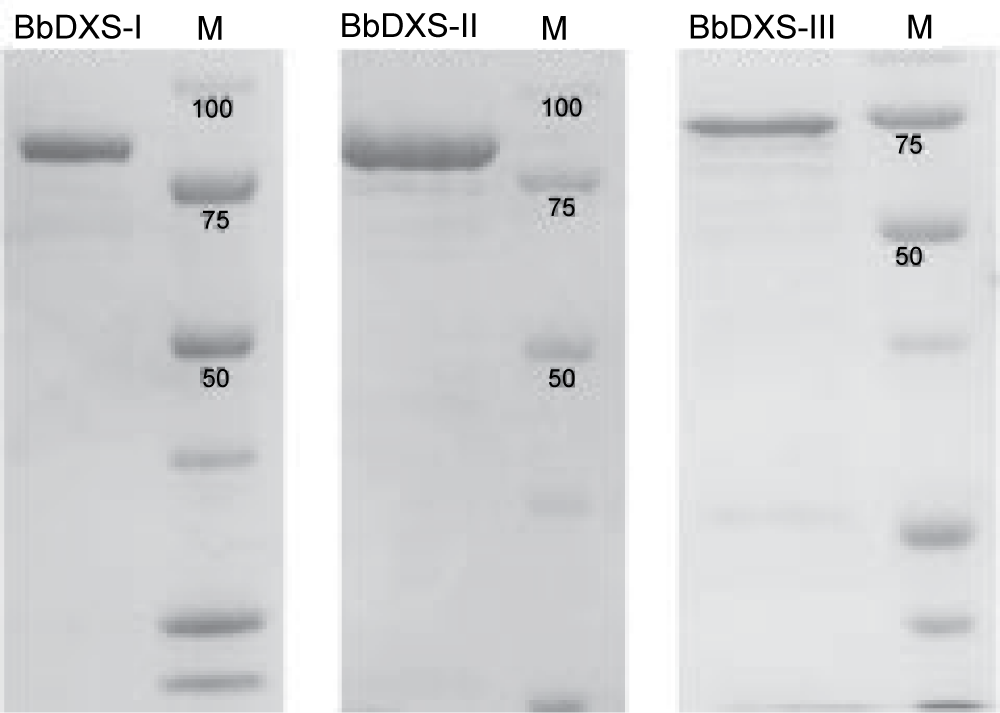
Fig. 11. SDS polyacrylamide gels of the purified DXS isoenzymes of B. braunii (stained with Coomassie Blue, M: marker)
The kinetic analyses of BbDXS-I, BbDXS-II and BbDXS-III gave hyperbolic saturation curves, and the Michaelis constants were determined by fitting hyperbolic plots of the initial reaction velocities versus substrate concentrations (see Fig. 12). Interestingly, we found considerable differences in the temperature dependence of enzyme activity among the three isoenzymes. BbDXS-III was much more stable at higher temperatures. Even at 50°C it maintained more than 70% of its maximum activity measured at 34°C (see Fig. 13).
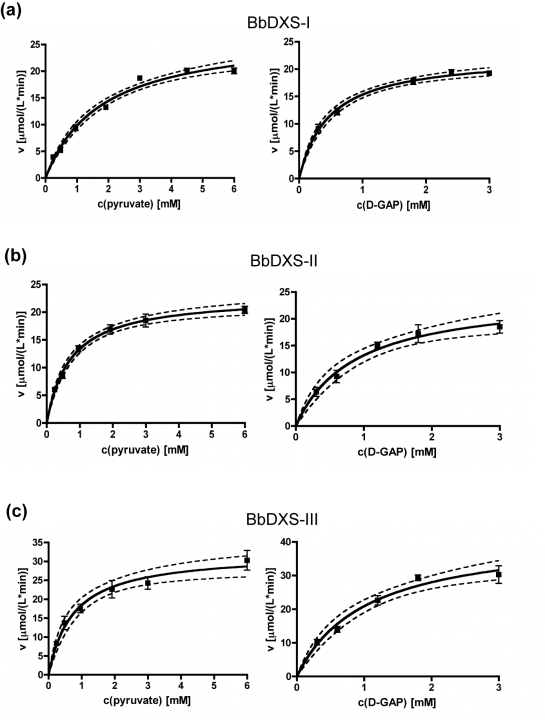
Figure 12. Substrate saturation curves of the three DXS isoenzymes. Dashed lines show the 95% confidence bands.
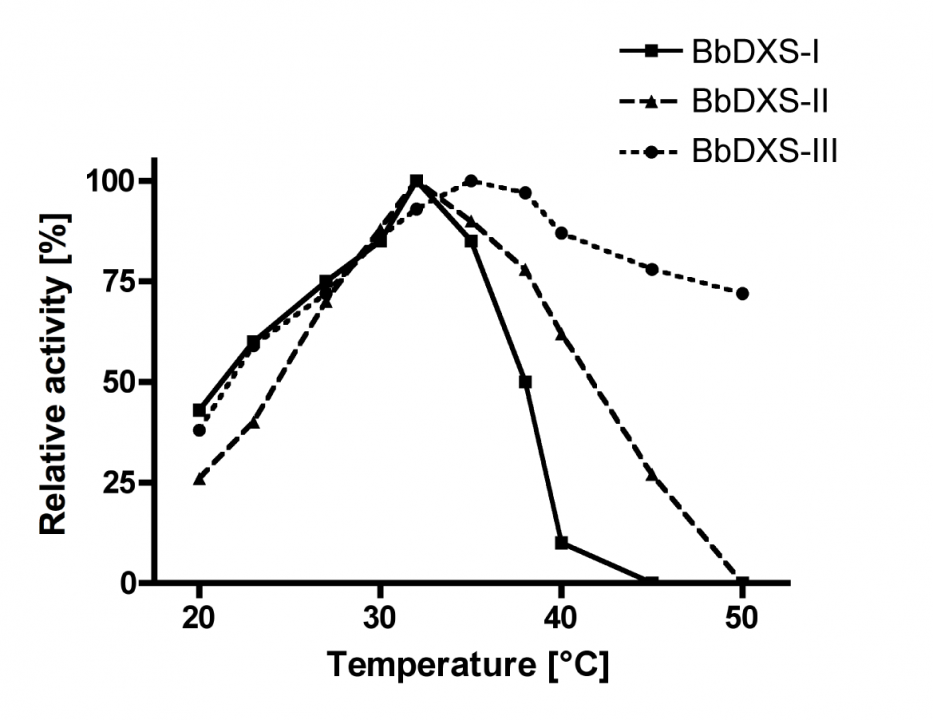
Figure 13. Temperature dependence of the relative activities of the three DXS isoenzymes.
Further details about the characterisation of the three B. braunii DXS isoenzymes can be found in Matsushima et al., 2012 (see publication list in section 4.1).
This was the first time that a green microalgae was demonstrated to have isoforms of DXS, which raises several questions. We still do not know whether this is a unique characteristic of B. braunii or of a higher taxonomic rank within the phylum Chlorophyta. From a biotechnological point of view, it will be interesting to learn more about the biochemical implications. The presence of DXS isoforms generates the potential to increase the metabolic flow through the pathway, but another function could be the selective utilisation under varying environmental conditions. We are planning to study the expression patterns of the isoforms in more detail. Further, we will be continuing characterisation of the other enzymes involved in botryococcene biosynthesis.
3.4 Effects of the Kaichū-Dōro causeway on biodiversity and environmental chemodiversity
(Collaboration with Dr. James D. Reimer, Associate Professor, University of the Ryukyus)
The Kaichū-Dōro causeway connects the Katsuren Peninsula on the east coast of Okinawa main island to the Henza island. The Japanese term kaichū (海中 , かいちゅう) means “in the sea”, and the word dōro (道路 , どうろ) means “road” or “highway”. Therefore, Kaichū-Dōro can be simply translated as “road in the sea”. Effectively, it is a 5 km long dyke or causeway with two flow-through channels (see Fig. 14). It was build in 1971 and thus has been in use for about 40 years.

Figure 14. Aerial photograph of Kaichū-Dōro connecting the main island of Okinawa and Henza island. The numbered circles depict sampling sites along the causeway. (Courtesy of Dr. J. D. Reimer)
The causeway cuts the Kin Bay area into separate habitats, and already at first sight, one can easily observe differences in vegetation and substrate. For instance, the north side is dominated by seagrass beds, while the the south side of the road has much more mud. The overall concept is to examine the biodiversity of many taxa including fish, echinoderms, amphipods and microorganisms throughout a one-year time course in order to assess the effects of Kaichū-Dōro on biodiversity. In this combined project, our role is to examine the microbiological diversity for all three domains of life, i.e. bacteria, archaea and eukaryotes. We are using “next generation” amplicon sequencing techniques and a fingerprinting method called “Denaturing gradient gel electrophoresis” (DGGE, see Fig. 15) to analyse three types of samples: (1) seawater, (2) low-tide sediment and (3) high-tide sediment.
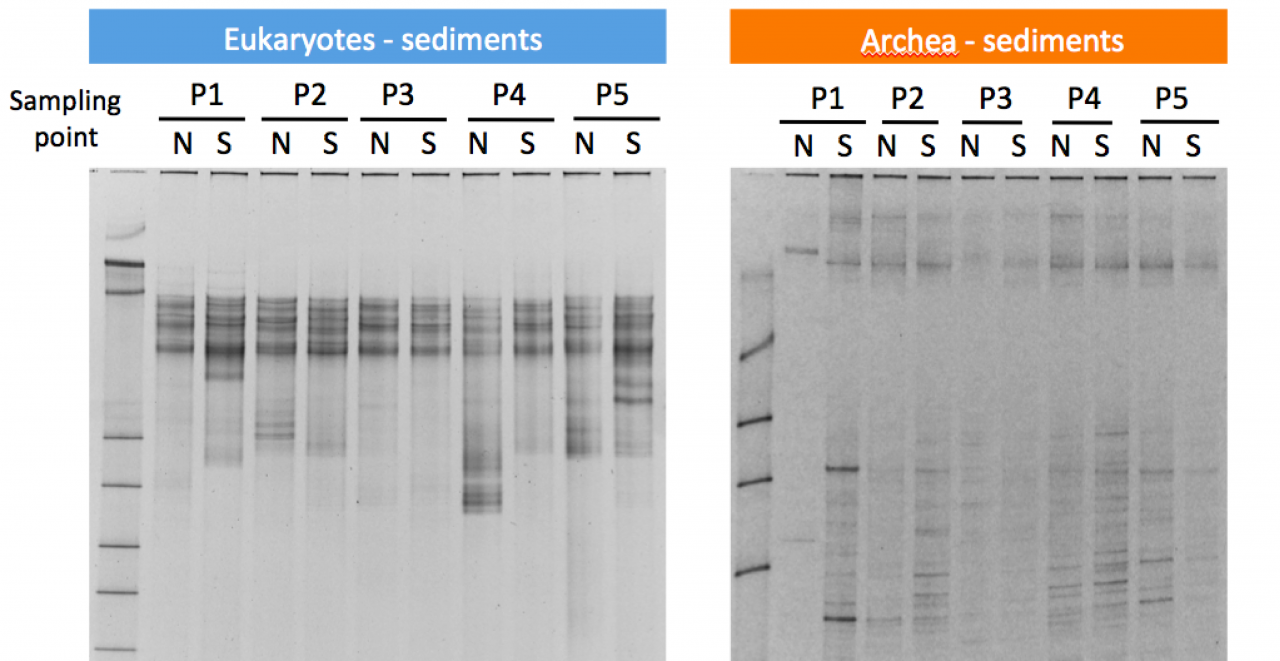
Figure 15. Examples of DGGE analysis of sediment samples. This method separates DNA according to differences in their base sequence. Differences in the band pattern reveal variations of the community structures. The labels P1-P5 refer to the sampling sites (see Fig. 12). The labels N and S indicate the north and south side of the causeway.
The first sampling took place in November 2011; the second sampling in February 2012. To get a complete seasonal cycle, further samplings are projected to be held in April 2012 and August 2012. We are now still in the process of sampling and data generation, and the main analyses are expected to be finished at the beginning of FY 2014. However, preliminary results for various taxa confirm many variations between the north and south side of the road. Amplicon data for eukaryotic microorganisms show pronounced differences (see Fig. 16).
![]()
Figure 16. Taxonomic composition (class level) at different sampling spots as inferred by high-throughput amplicon sequencing. Different colours depict different classes of eukaryotic microorganisms. Purple: Bacillariophyceae, dark blue: Agaricomyces, light blue: Glomeromycetes, green: no rank.
The same is true for the bacterial communities. When comparing the seawater of the north and south side of sampling point 1, which is the one closest to the main island, there is a striking difference with regard to the cyanobacterial portion (see Fig. 17). Furthermore, the southern water was extremely dominated by gamma-proteobacteria, which was caused by the fact that a single genus of the family Enterobacteriaceae amounted to more than 50% of the sequence reads.
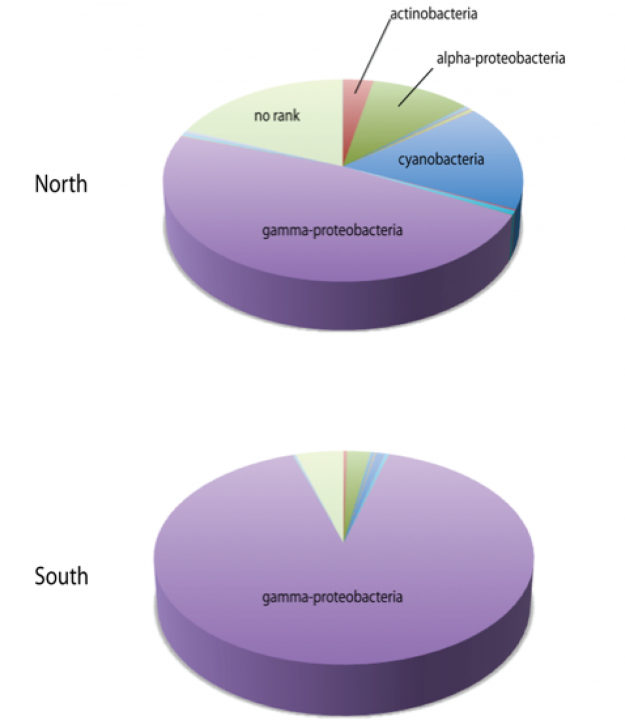
Figure 17. Bacterial composition of the seawater from the north and south side of sampling point 1 as inferred by high-throughput amplicon sequencing.
In addition to assessing the coastal biodiversity around Kaichū-Dōro, we are also interested in the environmental chemodiversity of that area. We have got sediment cores from all sampling sites, which will be used for geochemical dating analyses. These cores can be also used for screening special metabolites in order to correlate the distribution of sediment compounds with environmental factors and biodiversity data.
3.5 Secondary metabolites of the cyanobacterium Nostoc punctiforme and its symbiotic potential
Nostoc punctiforme is a multicellular, filament-forming cyanobacterium. It shows complex physiological and ecological features including nitrogen fixation, cell differentiation and symbiotic interactions with various plants and fungi [13]. From the genome sequence it is known that N. punctiforme has the genetic potential to biosynthesise several secondary metabolites belonging to the groups of polyketides (biosynthesised by polyketide synthases, PKS), nonribosomally made peptides (biosynthesised by nonribosomal peptide synthetases, NRPS) and hybrid structures thereof (biosynthesised by hybrid PKS/NRPS enzyme systems). In co-operation with the groups of Professor E. Dittmann (University of Potsdam, Germany) and Professor A. Liaimer (University of Tromsø, Norway) we could contribute to demonstrating that one of the polyketide compounds interferes with cellular differentiation events.
A bioinformatic survey for PKS and NRPS gene clusters revealed the presence of 11 clusters. Three of them are involved in glycolipid production and the biosynthesis of nostopeptolide and anabaenopeptin, respectively, while the compounds that are produced by the enzymes encoded by the other clusters are unknown. According to the bioinformatic analysis, the gene cluster that was named pks2 encodes for a hybrid enzyme system comprising seven PKS proteins with eight synthesis modules (as one of the proteins contains two modules) and one NRPS protein (see Fig. 18). Altogether the cluster spans around 63 kbp. The open reading frame (ORF) NpF3173, which encodes the NRPS module, was knocked-out by insertional mutagenesis, and the mutant obtained was analysed with regard to morphology and its behaviour in functional symbiosis assays.
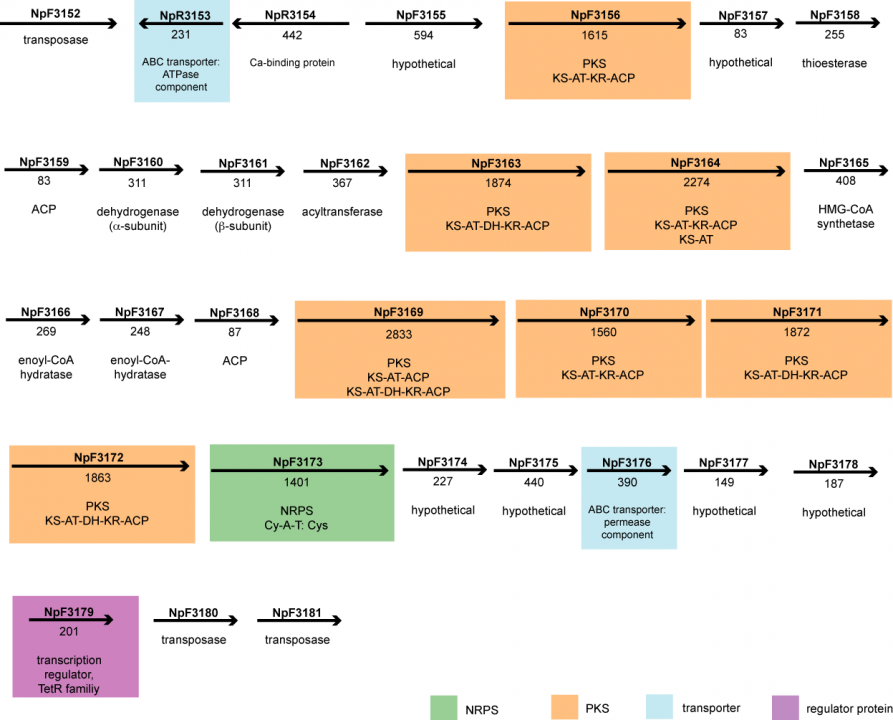
Figure 18. Schematic representation of the hybrid PKS/NRPS gene cluster pks2 identified in the genome of the cyanobacterium Nostoc punctiforme. The arrows depict the direction of transcription, and the numbers beneath the arrows indicate the number of amino acid residues encoded by the respective ORF.
Compared to the wild type, the mutant showed considerable changes in its cellular differentiation patterns. For instance, an accumulation of short filaments with terminal heterocysts (the cells that perform the fixation of molecular nitrogen) could be observed in liquid medium without combined nitrogen. Also, the mutant formed gliding filaments, so-called hormogonia, untypically over the entire growth curve and formed the akinete cell type, which is a kind of dormant cell with thick cell walls, much earlier than the wild type did under the same cultivation conditions. Further details about those results and a detailed discussion can be found in Liaimer et al., 2011 (see publication list in section 4.1).
Also, we initiated a new study on N. punctiforme to better understand the metabolic changes accompanied by the establishment of a symbiotic relationship. When forming symbiosis with plants, the cyanobacterium often locates in the roots. Since light cannot penetrate the root system, the bacterium must rely on organic carbon from the host for survival. N. punctiforme has the capability to grow heterotrophically in complete darkness using glucose, fructose or sucrose as a carbon and energy source [14]. This is in contrast to many other cyanobacteria, which inevitably need some light even if they are able to uptake sugars or alternative nutrients. The cultivation of N. punctiforme in the dark is a simple model to analyse its metabolism in the stage of symbiosis. The light state represents the free-living form, whilst the dark state mimics the symbiotic form. We have cultured the cyanobacterium with and without sugars, in the presence and absence of combined nitrogen, and in light and dark. In order to compare global gene expression patterns in light and dark states, we have initiated microarray experiments. These experiments will be continued in FY 2012.
Literature cited
[1] K.A. Steidinger, Some taxonomic and biologic aspects of toxic dinoflagellates, in: I.R. Falconer (Ed.) Algal toxins in seafood and drinking water, Academic Press, San Diego, 1993, pp. 1-28.
[2] J.D. Reimer, K. Takishita, S. Ono, T. Maruyama, Diversity and evolution in the zoanthid genus Palythoa utilizing nuclear ITS-rDNA, Coral Reefs, 26 (2007) 399-410.
[3] R.E. Moore, P.J. Scheuer, Palytoxin: a new marine toxin from a coelenterate, Science, 172 (1971) 495-498.
[4] P. Seemann, C. Gernert, S. Schmitt, D. Mebs, U. Hentschel, Detection of hemolytic bacteria from Palythoa caribaeorum (Cnidaria, Zoantharia) using a novel palytoxin-screening assay, Antonie Van Leeuwenhoek, 96 (2009) 405-411.
[5] P. Ciminiello, C. Dell'Aversano, E. Dello Iacovo, E. Fattorusso, M. Forino, L. Grauso, L. Tartaglione, F. Guerrini, R. Pistocchi, Complex palytoxin-like profile of Ostreopsis ovata. Identification of four new ovatoxins by high-resolution liquid chromatography/mass spectrometry, Rapid Commun Mass Spectrom, 24 (2010) 2735-2744.
[6] H.H. Senousy, G.W. Beakes, E. Hack, Phylogenetic placement of Botryococcus braunii (Trebouxiophyceae) and Botryococcus sudeticus isolate UTEX 2629 (Chlorophyceae), J Phycol, 40 (2004) 412-423.
[7] B.A. Knights, A.C. Brown, E. Conway, B.E. Middleditch, Hydrocarbons from the green form of the freshwater alga Botryococcus braunii, Phytochemistry, 9 (1970) 1317-1324.
[8] T.D. Niehaus, S. Okada, T.P. Devarenne, D.S. Watt, V. Sviripa, J. Chappell, Identification of unique mechanisms for triterpene biosynthesis in Botryococcus braunii, Proc Natl Acad Sci U S A, 108 (2011) 12260-12265.
[9] L.W. Hillen, G. Pollard, L.V. Wake, N. White, Hydrocracking of the oils of Botryococcus braunii to transport fuels, Biotechnol. Bioeng., 24 (1982) 193-205.
[10] M. Lohr, J. Schwender, J. Polle, Isoprenoid biosynthesis in eukaryotic phototrophs: a spotlight on algae, Plant Sci, (2011).
[11] M. Rodriguez-Concepcion, A. Boronat, Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics, Plant Physiol, 130 (2002) 1079-1089.
[12] E. Cordoba, H. Porta, A. Arroyo, C. San Roman, L. Medina, M. Rodriguez-Concepcion, P. Leon, Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize, J Exp Bot, 62 (2011) 2023-2038.
[13] J.C. Meeks, E.L. Campbell, M.L. Summers, F.C. Wong, Cellular differentiation in the cyanobacterium Nostoc punctiforme, Arch Microbiol, 178 (2002) 395-403.
[14] D.S. Hoare, L.O. Ingram, E.L. Thurston, R. Walkup, Dark heterotrophic growth of an endophytic blue-green alga, Arch Microbiol, 78 (1971) 310-321.
4. Publications
4.1 Journals
Matsushima*, D., H. Jenke-Kodama*, Y. Sato, Y. Fukunaga, K. Sumimoto, T. Kuzuyama, S. Matsunaga and S. Okada (2012). The single cellular green microalga Botryococcus braunii, race B possesses three distinct 1-deoxy-D-xylulose 5-phosphate synthases. Plant Sci 185: 309-320. (*these authors contributed equally) [Available online 10th January 2012, peer-reviewed]
Liaimer, A., H. Jenke-Kodama, K. Ishida, K. Hinrichs, J. Stangeland, C. Hertweck and E. Dittmann (2011). A polyketide interferes with cellular differentiation in the symbiotic cyanobacterium Nostoc punctiforme. Env Microb Rep 3: 550-558. [Published October 2011, peer-reviewed]
Note: The following publication is co-authored by a unit member, but is based on work that was mainly performed at another institution:
Sato, S., T. Nishimura, K. Uehara, H. Sakanari, W. Tawong, N. Hariganeya, K. Smith, L. Rhodes, T. Yasumoto, Y. Taira, S. Suda, H. Yamaguchi and M. Adachi (2011). Phylogeography of Ostreopsis along West Pacific coast, with special reference to a novel clade from Japan. PLoS One 6(12): e27983. [Available online 2nd December 2011, peer-reviewed]
4.2 Books and other one-time publications
Nothing to report.
4.3 Oral and Poster Presentations
H. Jenke-Kodama, J. D. Reimer, M. Tamura, Y. Taira, D. Richter. Analysis of toxin distribution patterns in Palythoa tuberculosa by integrated genomics. VII. US-Japan Seminar on cross-disciplinary expansions in marine bioorganic chemistry, 12th - 16th December 2011, Ginowan, Okinawa. [Poster presentation]
Y. Taira, M. Tamura, J. D. Reimer, H. Jenke-Kodama. Marine chemistry at higher stage: Analysis of toxin distribution patterns in Palythoa tuberculosa by integrated genomics. Rising Star Workshop "Advances in tropical island studies", 8th December 2011, Ryukyu University, Nishihara, Okinawa. [Poster presentation]
M. Tamura, J. D. Reimer, H. Jenke-Kodama. Marine chemistry at higher stage: Analysis of toxin distribution patterns in Palythoa tuberculosa by integrated genomics. Jacques Monod Conference "Integrated ecological genomics", 15th - 19th October 2011, Roscoff, France. [Poster presentation]
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report.
6. Meetings and Events
6.1 Seminar
- Date: 22nd April 2011
- Venue: OIST Campus Lab 1
- Speaker: Professor Takeo Horiguchi, Faculty of Science, Hokkaido University
- Title: "Evolution of Chloroplasts in Dinoflagellates - Tertiary Endosymbiosis and Kleptochloroplasts"
- _______________________________________________________________________________________
- Date: 5th March 2012
- Venue: OIST Campus Lab 1
- Speaker: Professor Shigeru Okada, Graduate School of Agricultural and Life Sciences, The University of Tokyo
- Title: "Let's make petroleum in your pond! Hydrocarbon production by the green microalga Botryococcus braunii"
7. Other
External funding gained in FY 2011:
- Resona Foundation. Project title: Assessing biodiversity along the Kaichū-Dōro leeway in Okinawa; 980,000 JPY (Principal Investigator: James D. Reimer, University of the Ryukyus; Co-Investigators: Maiko Tamura, Holger Jenke-Kodama, Kristine White, Masami Obuchi and Kazunori Tachihara)
- Nissei Foundation. Project title: Assessing the long-term impact of a leeway on coastal biodiversity forty years after construction; 1,500,000 JPY (Principal Investigator: James D. Reimer, University of the Ryukyus; Co-Investigators: Masami Obuchi, Kristine White, Maiko Tamura, Holger Jenke-Kodama, Kazunori Tachihara)



