FY2019 Annual Report
Protein Engineering and Evolution Unit
Assistant Professor Paola Laurino

Abstract
In the past few decades protein engineering allowed to generating artificial enzymes able to catalize unnatural reactions but also to become importnat tools for synthetic biology. Our Unit is particulary interested in generating new enzymes by directed evolution but also to understand how enzymes evolve in Nature. In FY19 we focused most of our attention on enzymes promiscuities and its origins. We also started few new exciting projects on phenotipic mutations, reconstruction of primordial folds and generation of active materials.
1. Staff
- Dr. Madhuri Gade, Researcher
- Dr. Babak Bahkshinejad, Researcher (till October)
- Dr. Bhanu Chouhan, Researcher
- Dr. Mirco Dindo, Researcher (JSPS Fellow from October)
- Dr. Saacnicteh Toledo Patino, Researcher (from December)
- Dr. Benjamin Clifton, Researcher (from January)
- Ms. Shayida Maimaiti, Technician (till April)
- Dr. Gen-ichiro Uechi, Technician
- Ms. Desirae Martinez, Technician (from July)
- Ms. Dalmira Merzhakupova, Graduate student (till May)
- Mr. Stefano Pascarelli, Graduate Student (from May)
- Mr. Dan Kozome, Graduate Student (from April to August)
- Mr. Andrea Testa, JSPS strategic program fellow (from February)
- Ms. Angela Kirykowicz, Graduate Student (till April)
- Ms. Upasana Pal, Research Intern (from April to May)
- Ms. Varsha Venkatesha Murthy, Research Intern (from April to May)
- Mr. Tatsuya Matsuda, Research Intern (till April)
- Ms. Nonno Hasegawa, Research Intern (from April till August)
- Ms. Catalina Mosquera Salcedo, Research Intern (from July till November)
- Ms. Anna Magdalena Klarkowska, Research Intern (from October)
- Ms. Sachie Matsuoka, Research Unit Administor
2. Collaborations
2.1 Topic: Discovery of a new metabolite in human cells towards investigation of human methionine adenosyltransferases promiscuities
- Type of collaboration: Joint research
- Researchers:
- Professor Colin Jackson, ANU, Australia
2.2. Topic: Protein Evolution after genome duplication: case study of EGFR in fish lineage
- Type of collaboration: Joint research
- Researchers:
- Professor Federica Di Palma, Earlham Institute, UK
2.3 Topic: Are epistatic patches in Rossmann Fold enzymes conserved?
- Type of collaboration: Joint research
- Researchers:
- Professor Michael Laessig, Cologne University, Germany
2.4 Topic: Enzymatically Active droplets
- Type of collaboration: Joint research
- Researchers:
- Professor Eric R. Dufresne, ETHZ, Switzerland
2.5 Topic: The functional instability of alanine:glyoxylate aminotransferase
- Type of collaboration: Joint research
- Researchers:
- Professor Barbara Cellini, Perugia University, Italy
- Professor Giorgio Giardina, Rome University, Italy
3. Activities and Findings
3.1 Discovery of a new metabolite in human cells towards investigation of human methionine adenosyltransferases promiscuities
Emerging powerful bioinformatics tools assign function to new protein based on structure, sequence and binding pocket similarities. However, these similarities do not invariably guarantee same specificity. Herein, we have assessed the relationship of identical binding pockets, similar structure and sequence of methionine adenosyltransferase (MAT) for different NTPs. MATs catalyze the formation of S-adenosyl-L-methionine (SAM) from ATP and methionine. We investigated at the biochemical, structural and cellular levels the specificities of the nucleotide binding pocket for NTPs in MAT enzymes from human (hMAT2A) and E. coli (eMAT). Our data shows that hMAT2A is promiscuous for different nucleotides whereas eMAT is specific for the ATP. To unravel the structural basis for this promiscuity X-ray structures of eMAT and hMAT2A were solved. The crystal data show that nucleoside acts as handle for NTPs and it allows a productive binding in hMAT2A whereas NTPs are unstable in eMAT binding pocket. hMAT2A contributes to the promiscuities by a high level of flexibility in the binding pocket. For the first time the promiscuity of hMAT2A is reported to be relevant in vivo by identifying a new metabolite in human liver cells. The difference in specificities reported here suggest that peculiar evolutionary path of enzymes might be driven by intracellular metabolites concentrations.
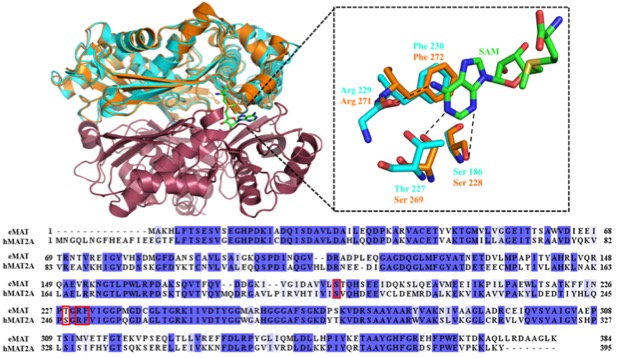
Figure 1: Structural and sequence aligment of MATs.
3.2 Study on the unusual evolutionary trajectory of Methionine adenosyltransferases
The third image, Figure 3, is a single image with the caption below it.
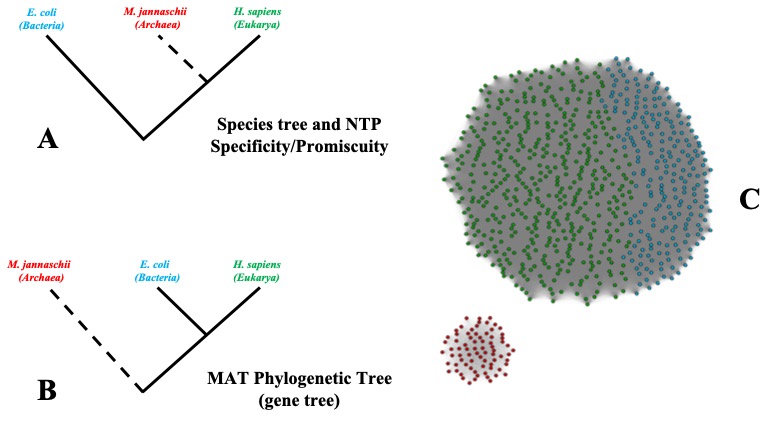
Figure 2: Schematic representation of observed trends in MAT.
3.3 Epidermal Growth Factor single mutants highlighted by homologs cross-conservation approach differentially affect cellular phenotype
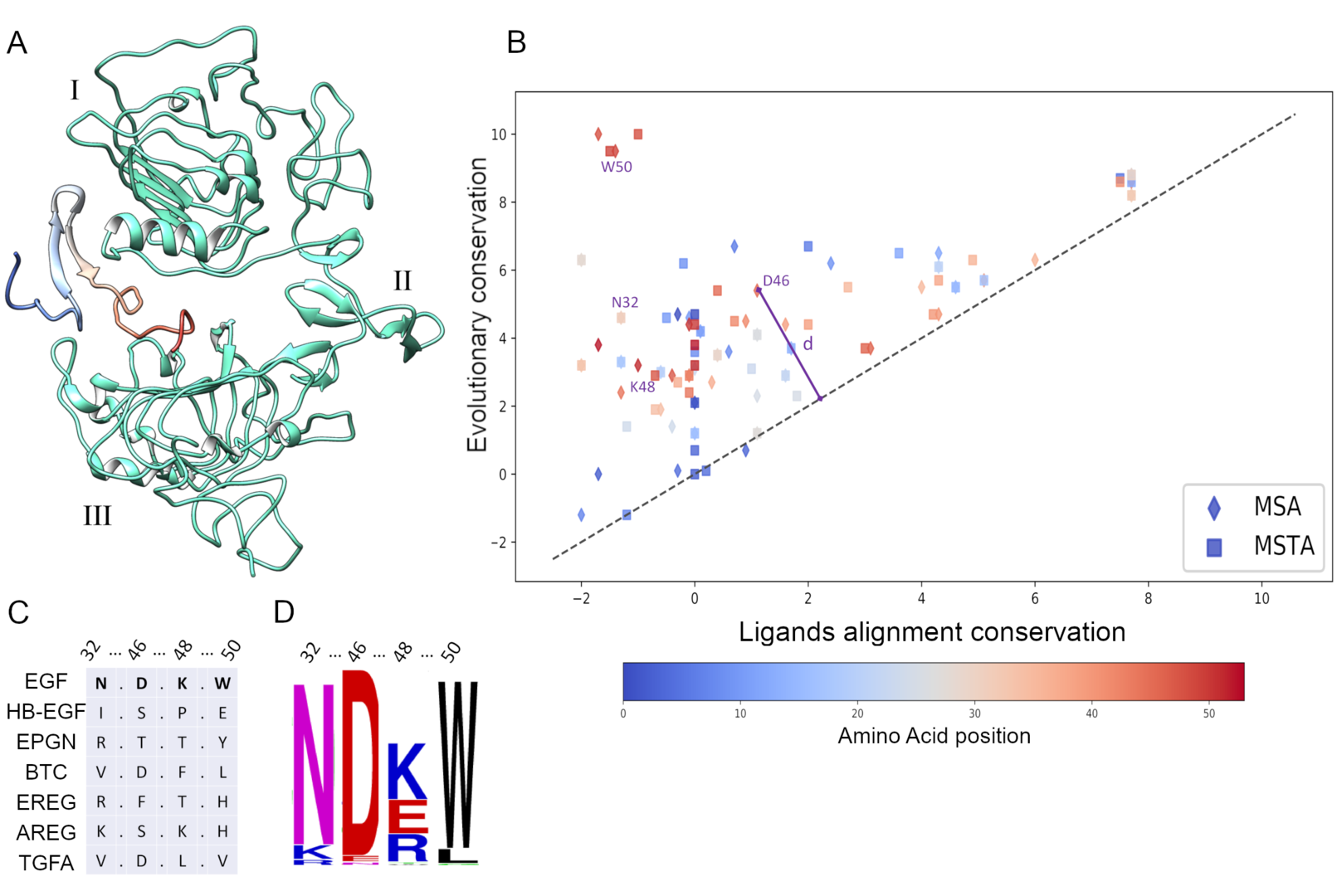
Figure 3: Cross-conservation analysis
4. Publications
4.1 Journals
Dindo, M., Grottelli, S., Annunziato, G., Giardina, G., Pieroni, M., Pampalone, G., Faccini, A., Cutruzzola, F., Laurino, P., Costantino, G. & Cellini, B. Cycloserine Enantiomers Are Reversible Inhibitors of Human Alanine:Glyoxylate Aminotransferase: Implications for Primary Hyperoxaluria Type 1. Biochem J, doi:10.1042/BCJ20190507 (2019).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
Conference Poster Presentations
- Gade, M., Li, L. T., Briones, A. V., Jackson, C. J. & Laurino, P. Understanding enzyme specificity as a tool for cofactor engineering, Stonehill College, Boston US (2019).
- Gade, M., Li, L. T., Briones, A. V., Jackson, C. J. & Laurino, P. Understanding enzyme specificity as a tool for cofactor engineering, Vancouver, Canada (2019).
- Pascarelli, S., Merzakupova, D. & Laurino, P. Evolution of constrained systems: EGFR and its ligands explored by cross-conservation and gene duplication., Nove Hrady, Czech Republic (2019).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar: Making an artificial cell capable of Darwinian evolution and artificial cell techniques for evolutionary engineering
- Date: April 23, 2019
- Venue: C016, Lab1
- Speaker: Prof. Norikazu Ichihashi, the Unviersity of Tokyo
6.2 Seminar: Generation of de novo designed protein structure library
- Date: July 02, 2019
- Venue: C016, Lab1
- Speaker: Prof. Nobuyasu Koga, Institute for Molecular Science
6.3 Seminar: Connecting biology and electronics with artificial protein switches
- Date: July 08, 2019
- Venue: C209, Centre Bldg.
- Speaker: Prof. Kirill Alexandrov, Queensland University of Technology
6.4 Seminar: New Trends in D-Amino acid Biocatalysis
- Date: September 17, 2019
- Venue: C016, Lab1
- Speaker: Prof. Loredano Pollegioni, University of Insubria
6.5 Seminar: Proteins from Peptides
- Date: January 31, 2020
- Venue: C016, Lab1
- Speaker: Prof. Andrei N. Lupas, Max-Planck Institute for Developmental Biology
7. Other
Internal Seminar Series
- Date: July 27, 2019 (16:30-17:00)
- Title: Understanding enzyme specificity as a tool for cofactor engineering
- Speaker: Dr. Madhuri Gade
Okinawa Science Mentor Program (OSMP2019)
- Period: July 26 - August 23, 2019
- Mentor for high school student: Mr. Stefano Pascarelli
Community Awards (PCD Group Project Class of 2018)
- Date: February 14, 2020
- Project Name: OldIST: Save Okinawans memory
- Awardee: Mr. Stefano Pascarelli
Researcher Appreciation Week (RAW!)
1. Oral presentation
- Date: March 02, 2020 (11:30am-)
- Venu: B250, Center bldg
- Presenter: Dr. Saacnicteh Toledo Patino
- Title: "Protein (r)evolution. Mimicking Nature to engineer artificial enzymes"
2. Poster presentation
- Date: March 05, 2020 (16:00)
- Venue: C700, Lab3
- Presenter: Dr. Saacnicteh Toledo Patino
- Title: "Protein LEGO. The puzzles of Nature"



