Annual Report FY2015: Reina Komiya
Reina Komiya
Science and Technology Associate
1. Introduction
Sexual reproduction transmits genetic information to the next generation and increases the genetic diversity of offspring. Successful sexual reproduction in flowering plants depends on accurate germ cell differentiation.Molecular mechanisms of germ cell development during pre-meiotic stages remain unknown in plants.
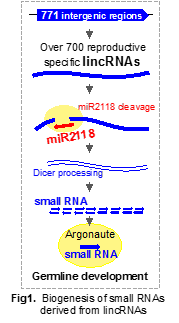
I have identified over 700 types of long intergenic non-coding RNAs (lincRNAs) specifically expressed during rice reproductive stages in which germ cell differentiation occurs. Furthermore, lincRNAs that contain the consensus sequence complementary to microRNA 2118 (miR2118) are cleaved within the miR2118 site. Cleaved lincRNAs are processed via DICER-LIKE4 (DCL4) protein, resulting in production of 21-nucleotides small RNAs (Figure 1, Komiya et al., 2014).
Non-coding RNAs (ncRNAs) are currently an important topic in biology.More than 90% of the genomes of higher organisms comprise intergenic regions. In many organisms, large numbers of endogenous ncRNAs play important roles at various developmental stages.However, most of ncRNA functions, transcriptional mechanisms, and interacting molecules of ncRNAs remain unknown in plants.
To discover reproductive mechanisms specific to pre-meiotic stages, I am engaged in two main projects using rice:
1. Determining the roles of over 700 lincRNAs, miR2118 and 21-nt small RNAs in early reproduction
2. Chromatin regulation by CRF between germ cells and somatic cells
By integrating 1 and 2, my goal is to construct an RNA/chromatin network model in pre-meiotic germ cells, including the topological organization of ncRNA, multichromosomal regions and chromatin regulators.
2. Activities and Findings
2.1 Roles of over 700 reproductive lincRNAs and miR2118
To reveal the function of lincRNAs and miR2118, the targeting system that causes deletion at specific targeting loci was used. Presently, we got the following 4 types of gene-targeting plants.
- single lincRNA mutants
- multiple lincRNAs mutants
- miR2118 mutants
- lincRNAs super-cluster mutants
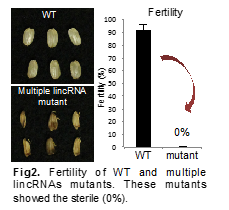
Single lincRNA mutants (1), in which 47bp was deleted, showed the fertility likely to wild-type plants. In contrast, multiple targeting mutants (2) showed the sterility from 0% to 2.6% (Figure 2). These results suggest that multiple lincRNAs may regulate early reproduction, though single lincRNA doesn’t. We also analyze T1 generations of miR2118 and lincRNAs super-cluster mutants (3, 4).
2.2 Transcriptional regulation of reproductive lincRNAs
To reveal epigenetic transcription of over 700 lincRNAs, I performed DNA methylation analysis genome-widely using leaf and anther, in which lincRNAs are expressed specifically. In informatics of DNA methylation, over 2000 hypo-methylated regions during reproduction were detected in the rice genome (Komiya unpublished data). However, most of these differentially DNA methylated regions are annotated at promoters of genes, not lincRNAs. Furthermore, DNA-FISH analysis showed that there was no specific localization of non-coding regions, where reproductive lincRNAs are expressed, at rice germ cells. These results suggest that epigenetic regulation and chromosome dynamics may not cause lincRNAs transcription.
2.3 Biological role of Chromatin Regulation Factor (CRF) in rice
CRF is a transcriptional repressor that directly binds to the modified histone for transcriptionally silenced heterochromatin in animals (Bannister and Kouzarides, 2005). Rice crf mutants showed sterility. Furthermore, some lincRNAs expressions are suppressed in crf mutants. These results suggest that CRF is required for development of rice reproductive tissues (Komiya unpublished data). Presently we made the CRF promoter: CRF:GFP transgenic plants, so I plan to reveal the biological roles of CRF by detecting CRF DNA binding sites using chromatin immuno-purification in FY2016.
3. Collaborations
3.1 Dr. Tu N. Le, Japan Advanced Institute of Science and Technology (JAIST). Informatic analysis of DNA methylation is performed in conjunction with Dr. Tu N. Le.
3.2 Dr. Masaki Endo and Ms. Masahiro Mikami, National Institute of Agrobiological Sciences (NIAS). A CRISPER CAS9 vector is provided by Dr. Endo
3.3 Dr. Shohei Takuno, (SOKENDAI)
Sequences of all types of transposable elements, which are used for DNA methylation analysis, are provided by Dr. Takuno.
4. Publications and other output
4.1 Conference Presentations
Komiya, R. “The biogenesis of small RNAs derived from over 700 reproductive lincRNAs”. The 79th annual meeting of the Botanical Society of Japan, Niigata Japan, September (2015).
4.2 Conference Organization
Symposium Title 「Diverse physiological functions of plant non-coding RNA」
Symposium Organizers Ohtani, M. and Komiya, R.
The 79th annual meeting of the Botanical Society of Japan, Niigata Japan, September (2015).
4.3 Publications
Komiya, R*. Plant “Co-suppression” leading to RNAi. Non-coding RNA Text Book (Jikken Igaku). 26-27 (2015). (* Corresponding author)
5. External Funding
5.1 JSPS Young Scientist (B) PI: Komiya, R. 2014, April ~ 2017, March.
5.2 JSPS Innovative area (RNA Taxonomy) PI: Komiya, R. 2015, April ~ 2017, March.2016 January~July. Maternity leave & Child care leave