FY2013 Annual Report
Mathematical Biology Unit
Associate Professor Robert Sinclair

Abstract
The Mathematical Biology Unit figuratively returned to life this year, with the arrival of two new Researchers, and the exciting and varied research activity which followed. We have been very active, working in a number of fields, covering topics as diverse as dinosaur bone classification, fish genomics and the possible role of temperature variation in the spread of invasive species. What has been most exciting has been the many points at which mathematics and biology have intersected in productive and unexpected ways. For example, beautiful genomic analyses performed by Dr. Jun Inoue and his collaborators are driving new types of mathematical analyses of ancient whole genome duplications. I must repeat that it has been a delight to have unit members again, and Haozhe Zhang, despite the short time he was here as an intern, made a significant contribution to the positive and productive atmosphere. In keeping with the Unit's philosophy, we continue to engage in purely biological and purely mathematical work, in the strong belief that this will provide firm anchors for our challenging interdisciplinary work.
1. Staff
- Dr. Robert Sinclair, Associate Professor
- Dr. Jun Inoue, Researcher (from April 2013)
- Dr. Harshana Rajakaruna, Researcher (from December 2013)
- Mr. Haozhe Zhang, Research Intern (July - September, 2013)
- Ms. Shino Fibbs, Research Administrator
2. Collaborations
- Theme: Palaeontology
- Researchers:
- Senior Curator Thomas Rich, Museum Victoria (Australia)
- Prof. Patricia Vickers-Rich, Monash University (Australia)
- Researchers:
- Theme: Theoretical Virology
- Researchers:
- Professor Dennis Bamford, University of Helsinki
- Dr. Janne Ravantti, University of Helsinki
- Researchers:
- Theme: Integrable Systems
- Researchers:
- Professor Martin Guest, Waseda University
- Associate Professor Takashi Sakai, Tokyo Metropolitan University
- Researchers:
- Theme: Yeast Introns
- Researcher:
- Associate Professor Anthony Poole, University of Canterbury (New Zealand)
- Researcher:
- Theme: Genome Evolution
- Type of collaboration: JSPS Grant 24770070
- Researchers:
- Professor Mutsumi Nishida, University of the Ryukyus
- Assistant Professor Yukuto Sato, Tohoku University
- Theme: Copepod Species Richness
- Researcher:
- Professor Mark Lewis, University of Alberta (Canada)
- Researcher:
3. Activities and Findings
3.1 Robert Sinclair
I have continued to work on projects relating to palaeontology, virology, introns, the combinatorics of sequence data and integrable systems. My main focus was work with Thomas Rich on a statistical analysis of the shape of a single arm bone (ulna) found in Australia some time ago. It has been a source of controversy because it seems to belong to a dinosaur group which is not thought to have spread to the Southern Hemisphere (Gondwana). Most of the year was spent on the manuscript describing this work. As a truly interdisciplinary project, it required quite a deal of effort to find descriptions and explanations which could be understood by researchers from such different fields as palaeontology and mathematics.
The work in virology is motivated by a capsid based classification of viruses, which has been put forward and investigated by Prof. Dennis Bamford over many years. One of the more technical issues is whether the sequences of proteins which have similar structures also show similarity of any kind. Standard bioinformatics tools have failed to detect similarity where it would be expected. My involvement began with the development of a software tool for more sensitive similarity detection, and continued all year with the application of this tool to a large data set. We made use of desktop machines and also the Tombo Computing Cluster. This work is ongoing.
In the work on yeast introns, the question is whether one can infer anything about the introns of an organism by examining its intronless genes. The idea is that intronless genes lack something - introns. This "lack" should be detectable by statistical analysis, and should in some sense appear as a footprint. This is interesting for a number of reasons. Mathematically, it is important because we do not have many tools for detecting the lack of something.
The work that I am doing with Prof. Guest of Waseda University is an attempt to better understand certain systems of differential equations which show unexpected structure. I suspect that these systems may have something to teach us about mathematical aspects of central pattern generators of animals. There is a possibility that one may be able to make a link between animals' reactions to certain stresses and the way that integrable systems lose their special properties in response to perturbation.
Finally, I have always been occupied with the relationship between mathematics and the natural sciences in general. My opinion article "Where Is Mathematics?" arose from my worry that the true nature of mathematics is misunderstood in a number of ways. In particular, I do not see mathematics as the basis of the physical world, although I am delighted to see how well mathematics and physics enrich each other, nor can I agree that mathematics can be seen as the unique product of the mind of Homo sapiens.
3.2 Jun Inoue: Evolutionary Genomics of Vertebrates
In order to understand the evolution of vertebrate genomes, we are focusing on the whole genome duplication (WGD) event which occurred just before the diversification of teleost fish (Figure 1, below) for the following reasons:
- Ancestral WGDs have occurred three times in vertebrate history, providing many surplus genetic resources for diversification.
- More evidence remains from the fish-specific WGD than from the other, older events for evolutionary study.
- A time-tree combining molecular and fossil evidence can be used to trace the teleost diversification pattern.
As a starting point for evolutionary study, we constructed a computational pipeline and carefully estimated the orthologs between tetrapods and teleosts for all approximately 20,000 protein coding genes. Based on this orthology information, we are working on the following two projects.
A. Temporal gene loss pattern: The orthology information allows us to estimate the gene loss/persistence pattern after WGD mathematically by counting the number of gene pairs derived from the fish-specific WGD (Fig. 1).
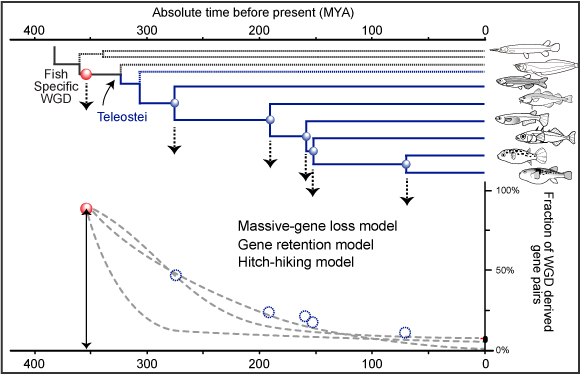
Figure 1.
B. Comparison at chromosomal level: The orthology information enables us to compare genomes at the chromosomal level (Figure 2, below).
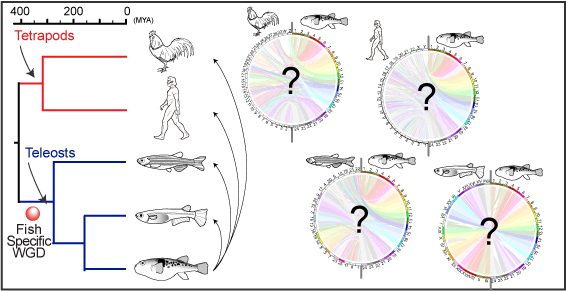
Figure 2.
3.3 Harshana Rajakaruna
Harshana has worked on the broad theme of how fluctuations of ocean temperatures may impact marine copepod species persistence and distribution from a theoretical and a mathematical modeling perspective, testing the models with empirical data. His work involved two major areas of research:
Does periodically forcing temperature reduce species taxonomic richness? The case specific to marine calanoid copepods.
Here, he worked on an extension to diversity distribution models in the metabolic theory of ecology and fitted them to new meta data collected from online interactive databases such as OBIS, and ESRL-NOAA. His work is in collaboration with Prof. Mark Lewis, Centre for Mathematical Biology, University of Alberta. The study showed that periodic fluctuations of temperature should reduce the effective metabolic rate of copepod species populations having short generation times, leading to lower species taxonomic richness. The new model may account for the effect of periodic fluctuations in species richness. The proposed extended MTE model may explain the macro-ecological patterns arising from periodic fluctuations of temperature on taxa subject to such variation, with short generation times, and suggests that temperature amplitude may contribute to both the longitudinal and the latitudinal differences in such species' taxonomic richness.
Does spatial variation in periodic temperature oscillations drive colonization of marine copepods?
Here, he worked on restructuring mathematical models dealing with the effect of temperature on the persistence and growth of marine copepod populations, and testing hypotheses, emerging from his models, using new meta data on marine copepod species' occurrence in the world's oceans, obtained through online databases such as OBIS and ESRL-NOAA. His work was done in collaboration with Prof. Mark Lewis, Centre for Mathematical Biology, University of Alberta. The study suggested that copepod populations migrating from regions with high to low amplitude temperature fluctuations on a yearly scale, having similar mean temperatures, have a greater potential to colonize, given other ecological and environmental factors are the same throughout the regions studied. Furthermore, based on 1 million records of species' occurrence data, comprising 645 marine calanoid copepod species, sampled at <150 m depth, covering world marine regions, he showed that the range of amplitudes of habitats that a species occupies may be determined by the maximum amplitude of temperature of the habitats in which the species is distributed. The results lead one to ask whether there exists a trend that marine copepods in high amplitude temperature habitats colonize low amplitude habitats within the same mean temperature range, increasing their potential to become invasive.
4. Publications
4.1 Journals
- Shiga, H. and R. Sinclair (2013). "The Space of 9-Dimensional Graded Algebras of Regular Type." Ryukyu Mathematical Journal 26: 61-69.
- Sinclair, R. (2014). "Where Is Mathematics? [Point of View]." Proceedings of the IEEE 102(1): 2-4.
4.2 Books and Other One-Time Publications
Nothing to report.
4.3 Oral and Poster Presentations
- Inoue, J. G. (2013). Tutorial of MCMCTREE Analysis. School of Life Sciences, Fudan University.
- Inoue, J. G., et al. (2013). 脊椎動物3億年のゲノム進化:ゲノム重複に由来する遺伝子の保持と進化に寄与した自然選択項の検出. University of Tsukuba.
- Inoue, J. G., et al. (2013). Selecting Molecular Markers from Transcriptome Data for Actinopterygian Phylogenetics. School of Life Sciences, Fudan University.
- J., I., et al. (2014). Selecting molecular markers from transcriptome data for fish phylogenetics. Workshop on Methods for biodiversity Research. Shanghai, China.
- Sinclair, R. (2014). Is Infinity Far Away? OIST Open Campus 2014.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar
Title: Fluctuating ocean temperatures that shape the global invasive marine species distribution.
- Date: June 5, 2013
- Venue: C016
- Speaker: Harshana Rajakaruna
- Centre for Mathematical Biology, Dept. of Biological Sciences, University of Alberta, Canada
6.2 Seminar
Title: Coelacanth Genome and Regulatory Evolution
- Date: June 17, 2013
- Venue: C209
- Speaker: Byrappa Venkatesh
- Professor and Research Director, Comparative Genomics Lab, Institute of Molecular and Cell Biology, A*STAR
6.3 Seminar
Title: Large data analysis and preliminary insights from human population genomics at Tohoku Medical Megabank Organization
- Date: March 28, 2014
- Venue: C016
- Speaker: Yukuto Sato
- Assistant Professor, Division of Biomedical Information Analysis, Dept. of Integrative Genomics, Tohoku Medical Megabank Organization, Tohoku University



