FY2018 Annual Report
G0 細胞ユニット
柳田充弘教授

Abstract
The G0 Cell Unit studies how cells in the proliferative (dividing) or quiescent (non-dividing) phase respond to nutritional shifts (e.g., nitrogen source starvation and glucose limitation). We identify gene products (mostly proteins) and chemical factors (small molecules, metabolites) that affect adaptation mechanisms in order to understand the basis for longevity of long-term quiescent cells. We employ fission yeast as a eukaryotic cell model as the great majority of this organism’s genes are conserved in human. In addition, we are studying chromosomal regulatory mechanisms involving condensin, cohesin complexes (which lead to proper chromosome segregation in proliferating cells), and other nutrient adaptation-related nuclear chromatin proteins. This line of studies aims to understand dynamics of nuclear chromatin in response to nutritional cues. Study on human blood metabolomics initiated from comparative study with fission yeast metabolomics now directly aims to identify and understand the roles of metabolites intimately related to human aging.
Our current principal research projects may be summarized as below:
(1a) Methodological development for metabolomic analysis.
(1b) Comprehensive and quantitative human metabolomics (blood, saliva, and urine).
(2) Understanding cell regulation in response to nitrogen deprivation.
(3) Understanding cell regulation in response to glucose starvation.
(4) Understanding chromosomal regulatory mechanisms involving condensin, cohesin complexes, and other nutrient adaptation-related nuclear proteins.
In FY2018, we published 7 original articles on research topics including chromosome regulatory mechanisms (separase, cohesin and DNA topoisomerase II), genetic regulation in G0 quiescent cells, and human metabolomic analysis, as listed in 3. Activities and Findings and 4. Publications.
1. Staff
- Dr. Yung-Ju Chen, Postdoctoral Scholar (until August)
- Dr. Takeshi Hayashi, Staff Scientist
- Dr. Susumu Morigasaki, Staff Scientist
- Dr. Norihiko Nakazawa, Staff Scientist
- Dr. Kenichi Sajiki, Staff Scientist
- Dr. Xingya Xu, Postdoctoral Scholar (until December)
- Dr. Haifeng Zhang, Postdoctoral Scholar
- Ms. Orie Arakawa, Technician
- Ms. Ayaka Mori, Technician
- Ms. Michiko Suma, Technician
- Ms. Yuria Tahara, Technician
- Ms. Junko Takada, Technician (until February)
- Dr. Takayuki Teruya, Technician
- Ms. Risa Uehara, Technician
- Ms. Li Wang, Technical staff (until December)
- Ms. Chikako Sugiyama, Research Unit Administrator
2. Collaborations
- Theme: Analysis of human blood metabolites, involved in aging and aging-related diseases
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Takumi Mikawa, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Comparative analysis of blood metabolome between healthy people and metabolic abnormality patients
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Masuzaki, Division of Endocrinology, Diabetes and Metabolism, Hematology, Rheumatology (Second Department of Internal Medicine), Graduate School of Medicine, University of the Ryukyus
- Theme: Blood metabolome analysis in abnormal cerebral function
- Type of collaboration: Joint research
- Researchers:
- Dr. Yasuhide Fukuji, Director, National Hospital Organization Ryukyu Hospital
- Dr. Taku Otsuru, Deputy Director, National Hospital Organization Ryukyu Hospital
3. Activities and Findings
Here, we introduced four research activities in G0 Cell Unit on FY2018.
3.1 Suppressor mutation analysis combined with 3D modeling explains cohesin’s capacity to hold and release DNA (From OIST news center article "Mutation and 3D Modelling Reveal New Structure to Cell Division Process" by Andrew Scott)
Cell division is one of the most fundamental processes of biology. It’s what allows all living things to grow, multiply and form complex structures. But scientists still don’t understand many of the details behind the process.
Before a cell divides, it makes two copies of each of its chromosomes. These copies, called sister chromatids, align – and segregate -- to opposite ends of nucleus so that a full set will accompany the two daughter cells that will form. In a new study published in Proceedings for the National Academy of Sciences (PNAS), researchers at Okinawa Institute of Science and Technology Graduate University (OIST) are challenging a key aspect of how this process has been assumed to work.
The study focuses on a protein complex called cohesin, which is tasked with holding the sister chromatids together until the cell begins to divide, and then letting go when the cell divides and the chromosomes must separate. Most researchers think that cohesin is shaped like a ring that holds chromatids together by encircling them. But for Drs. Xingya Xu and Norihiko Nakazawa at the OIST G0 Cell Unit headed by Professor Mitsuhiro Yanagida, some aspects of this model didn’t add up.
Cohesin releases its hold on chromatin with the help of an enzyme called Cut1. Chromatin is a mass of genetic material composed of DNA and proteins that condense to form chromosomes during eukaryotic cell division. When chromosomes are aligned, the Cut1 enzyme breaks open cohesin, allowing the two chromatids to segregate into the daughter cells. When the researchers mutated the gene encoding Cut1 in fission yeast cells, the cell division cycle broke down, as expected, because the mutated Cut1 couldn’t open the ring-shaped cohesin. However, in theory normal cell division could be restored by adding a second mutation that acts like an undo button for the first mutation, allowing the enzyme to break open cohesin once more, opening the ring.
That’s not what the team saw when they added this second mutation to their fission yeast cells. Instead, the cells that carried both these mutations were still not able to break down cohesin. Because the Cut1 enzyme needs cohesin’s assumed ring structure to function correctly, the scientists began to think that cohesin did not have a ring shape like researchers had long thought.
“We were surprised,” said Dr. Nakazawa, “because if cohesin was like a ring around chromatin, then the chromatids should have been cut loose by the second mutation.” The fact that it wasn’t, he adds, suggests that cohesin may have a different structure.
Previous studies in 2003 by Prof. Yanagida’s group had examined some aspects of cohesin’s structure at an atomic level using an extremely high-resolution microscope called an Atomic Force Microscope (AFM), but those results didn’t reveal all the fine details of the complex’s shape. To get a better picture, the OIST team had to look at the genetics.
The scientists first sequenced the whole genome of cohesion in a fission yeast cell to identify where mutations occurred in areas where proteins interact with it, and how these mutations affect the structure of the cohesin complex. The mutations arose in areas of cohesin known as the “head” and “hinge”, which fasten the two main units of the protein complex together. They are thought to be on opposite sides of the ring structure, but further genomic analysis of cohesin itself suggested that they might be located next to one another, which would make cohesin’s ring configuration unlikely.
Prof. Yanagida, Dr. Xu and Dr. Nakazawa proposed a new model, dubbed hold and release, in which cohesin is more like a jaw that holds chromatids in place and then opens to allow chromatin to move, without the need for Cut1 to break open the complex.
Based on the sequences of cohesin’s head and hinge, the researchers constructed a 3D computer model of the protein complex. The model, created by Dr. Ryuta Kanai and Prof. Chikashi Toyoshima at the University of Tokyo, showed that the second mutation in the OIST fission yeast study couldn’t break down cohesin – supporting their observations in fission yeast and adding more evidence that the complex was not ring-shaped.
The new model fit well with AFM images of cohesin from the older study, which had suggested that the molecule was half the length of the expected ring structure and that cohesin was “tadpole shaped” – large at one end and tapering down to a thinner, opposite end.
Dr. Nakazawa and Dr. Xu still don’t have enough evidence to pin down cohesin’s shape. They also still don’t know how cohesin binds to chromatin to hold it in place and whether a single complex or a pair of them is needed to do so. As the team explore these possibilities, they may get closer to proving their new theory – and perhaps even glean a better understanding of diseases caused by abnormal cell division.
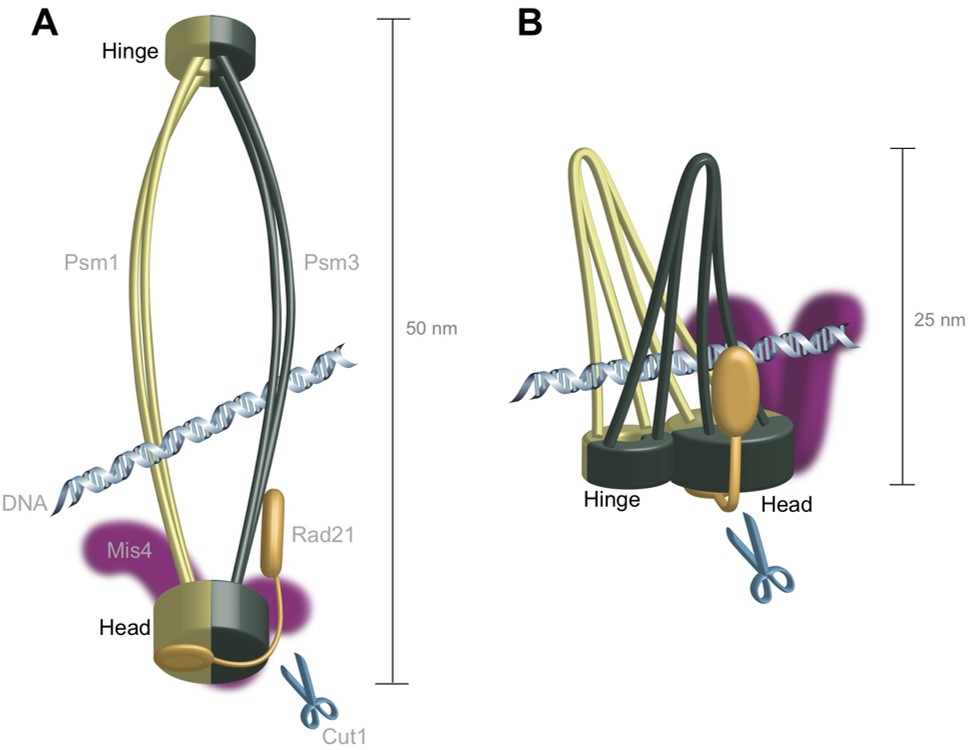
Figure1. These diagrams compare the two models of the cohesin protein’s structure. The ring model has distinct “head” and “hinge” features at opposite ends of the molecule, whereas the hold-and-release model suggests these features are located next to one another, which would mean the protein could hold DNA like a pair of pincers.
3.2 Genetic regulation of mitotic competence in G0 quiescent cells (From OIST news center article "The Secret Behind Cell Revival Revealed" by Ipsita Herlekar)
Scientists from the Okinawa Institute of Science and Technology Graduate University (OIST), have identified 85 genes essential for fission yeast cells at rest, under nutritionally limited environmental conditions to maintain their ability to go back to the dividing mode. The study sheds light on the genetic network required for resting cells like cancer stem cells to reactivate and has potential applications in developing new therapies to treat cancer. The study has been published in the journal Science Advances.
In all living organisms, cells multiply when nutritional and environmental conditions are favorable for cell division. The availability of ample nutrition via a nitrogen source is one of the main deciding factors necessary to initiate the cell division process, as nitrogen is needed to make DNA, RNA and protein. When an external nitrogen-source is absent, cells must recycle the intracellular nitrogen source and remain in a non-dividing (quiescent) state, alternatively known as the G0 phase. Regarding carbon source (the energy source), the medium contains excess carbon energy source (glucose) so that metabolism of G0 cells in brain or in muscle is fully active. In the G0 phase, the cells change into shapes different from the dividing mode and they modify the way they work to cope with the scarcity of the nitrogen source, reactivating for division only when favorable and ample amount of nitrogen returns. This ability of cells to restart the cell division process is referred to as mitotic competence (MC).
“Understanding how G0 cells maintain their ability to revive themselves on experiencing favorable conditions is important,” says Dr. Kenichi Sajiki, a researcher in the laboratory of Professor Mitsuhiro Yanagida, and the lead author of the paper. For this study, he and his colleagues from OIST's G0 Cell Unit examined a collection of 3280 fission yeast deletion mutant cell strains that had one of their genes, or a part of it deleted.
Fission yeast cells were encouraged to grow and multiply by providing them with necessary nutrients. Their nitrogen supply was subsequently cut-off to induce G0 phase. Once the G0 phase was induced, the yeast cells were maintained in this state for as long as four weeks. At the end of this time, they were brought back to their dividing mode by resuming their nitrogen supply.
Naturally occurring or wild-type fission yeast cells can maintain MC while they are in the G0 phase effortlessly for over four weeks, even after their nitrogen supply had been cut short. However, 85 out of the total 3280 mutant yeast strains examined exhibited shorter periods of MC. The scientists therefore inferred these 85 missing genes to be essential for yeast cells to maintain MC during their G0 phase. To their surprise, nearly half of these genes were reportedly linked to cancer, implicating a tight connection between the maintenance of MC in G0 cells and malignant divisions in cancer cells. Recently, stem cells in a cancerous tumor were known to remain in the G0 phase while being exposed to the currently used cancer therapies that target dividing cells, and later reactivate themselves and begin dividing again. The study of these 85 genes might therefore suggest new therapies that target such cancer stem cells.
Mutant yeast cells with shortest MC had missing within them nem1, a gene related to the de-phosphorylation of proteins. De-phosphorylation is a process necessary for transfer of signals within a cell, which takes place by changing the activity of diverse proteins. On the other hand, cells devoid of genes responsible for remodeling of DNA structure and degradation of unnecessary materials inside cells exhibited longer durations of MC, which implicated that these functions were required for prolonging G0 phase. These observations have therefore helped identify the timings of the required cellular functions to maintain MC.
On losing MC, the cells lacking nem1 showcased abnormally shaped nuclei. This was a result of loss of control in the signaling pathway, responsible for degrading cell parts. Dr. Sajiki also found Nem1 to de-phosphorylate another gene product Ned1, which controls lipid balance inside cells, and dysfunction of which leads to disintegration of the nucleus. This therefore confirms the association of the two genes nem1- ned1, to play a significant role in protecting the nucleus during the cell’s transition from proliferation to quiescence, leading to excessive degradation of internal cell components while maintaining MC.
This relationship of nem1 as a regulator and ned1 as a target, also suggests a new framework of the genetic network to maintain MC, since supporting studies have showed nem1 was required for MC maintenance exclusively in the G0 phase, while ned1 was required both in the G0 and dividing phases. Some genes may be working as G0-specific switches, and their target genes may be working as housekeeping genes. The scientists named such genes for the G0 switches as G-zero essential (GZE) genes and genes for housekeeping as super housekeeping (SHK) genes and identified about 80 genes for each group.
Professor Yanagida says, “the paper is a real breakthrough for our understanding of how MC is maintained within G0 cells. Two groups of genes, each with less than 100 members, are needed for MC maintenance, and at least in one case Nem1 and Ned1, each representing two groups, directly interacted for protein dephosphorylation. Dr. Sajiki discovered the first group of SHK genes as a graduate student in OIST, and now, nine years later, he and his colleagues in the G0 Cell Unit at OIST have identified a new class of 85 GZE genes. I predict many exciting discoveries for Dr. Sajiki and other researchers interested in fully understanding the relationship of these genes.”
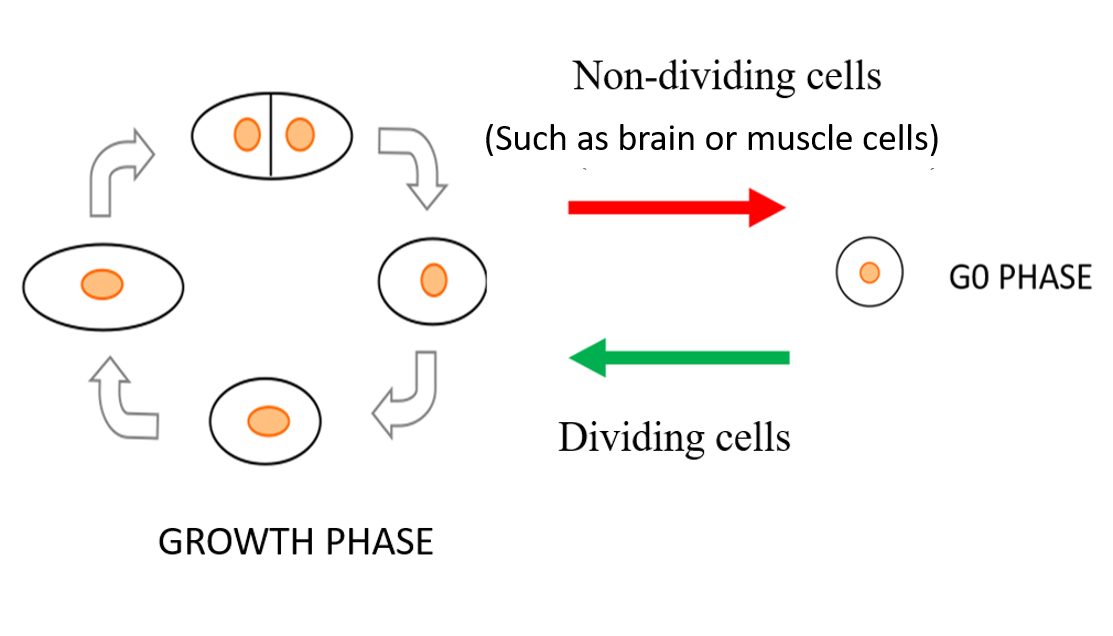
Figure 2. Under nutrition deficient conditions, fission yeast cells stop multiplying and slip into a resting state or the G0 phase. During G0 phase, the cells change shape and modify their functioning to survive under limited nitrogen availability and resuscitate only when nitrogen supplied conditions return.
3.3 Diverse metabolic reactions activated during 58-hr fasting are revealed by non-targeted metabolomic analysis of human blood (From OIST news center article "Fasting Ramps Up Human Metabolism, Study Shows" by Nicoletta Lanese)
Fasting may help people lose weight, but new research suggests going without food may also boost human metabolic activity, generate antioxidants, and help reverse some effects of aging. Scientists at the Okinawa Institute of Science and Technology Graduate University (OIST) and Kyoto University identified 30 previously-unreported substances whose quantity increases during fasting and indicate a variety of health benefits.
“We have been researching aging and metabolism for many years and decided to search for unknown health effects in human fasting,” said Dr. Takayuki Teruya, first author of the paper and a technician in the OIST G0 Cell Unit, led by Prof. Mitsuhiro Yanagida. “Contrary to the original expectation, it turned out that fasting induced metabolic activation rather actively.”
The study, published January 29, 2019 in Scientific Reports, presents an analysis of whole human blood, plasma, and red blood cells drawn from four fasting individuals. The researchers monitored changing levels of metabolites -- substances formed during the chemical processes that grant organisms energy and allow them to grow. The results revealed 44 metabolites, including 30 that were previously unrecognized, that increased universally among subjects between 1.5- to 60-fold within just 58 hours of fasting.
In previous research, the G0 Cell Unit identified various metabolites whose quantities decline with age, including three known as leucine, isoleucine, and ophthalmic acid. In fasting individuals, these metabolites increase in level, suggesting a mechanism by which fasting could help increase longevity.
“These are very important metabolites for maintenance of muscle and antioxidant activity, respectively,” said Teruya. “This result suggests the possibility of a rejuvenating effect by fasting, which was not known until now.”
Metabolites Give Clues to Mechanism and Health Effects
The human body tends to utilize carbohydrates for quick energy -- when they’re available. When starved of carbs, the body begins looting its alternate energy stores. The act of “energy substitution” leaves a trail of evidence, namely metabolites known as butyrates, carnitines, and branched-chain amino acids. These well-known markers of energy substitution have been shown to accumulate during fasting.
But fasting appears to elicit effects far beyond energy substitution. In their comprehensive analysis of human blood, the researchers noted both established fasting markers and many more. For example, they found a global increase in substances produced by the citric acid cycle, a process by which organisms release energy stored in the chemical bonds of carbohydrates, proteins and lipids. The marked increase suggests that, during fasting, the tiny powerhouses running every cell are thrown into overdrive.
Fasting also appeared to enhance the metabolism of purine and pyrimidine, chemical substances which play key roles in gene expression and protein synthesis. The finding suggests fasting may reprogram which proteins cells build at what time, thus altering their function. The change may promote homeostasis in cells, or serve to edit their gene expression in response to environmental influences.
When metabolized, purine and pyrimidine also boost the body’s production of antioxidants. Several antioxidants, such as ergothioneine and carnosine, were found to increase significantly over the 58-hour study period. Antioxidants serve to protect cells from free radicals produced during metabolism. Products of a metabolic pathway called the “pentose phosphate pathway” also stay the harmful effects of oxidation, and were similarly seen to increase during fasting, but only in plasma.
Newfound Health Benefits of Fasting?
The authors suggest that these antioxidative effects may stand as the body’s principal response to fasting, as starvation can foster a dangerously oxidative internal environment. Their exploratory study provides the first evidence of antioxidants as a fasting marker. In addition, the study introduces the novel notion that fasting might boost production of several age-related metabolites, abundant in young people, but depleted in old.
“Recent aging studies have shown that caloric restriction and fasting have a prolonging effect on lifespan in model animals...but the detailed mechanism has remained a mystery,” said Teruya. “It might be possible to verify the anti-aging effect from various viewpoints by developing exercise programs or drugs capable of causing the metabolic reaction similar to fasting.”
The findings expand on established ideas of what fasting could do for human health. The next step would be to replicate these results in a larger study, or investigate how the metabolic changes might be triggered by other means.
“People are interested in whether human beings can enjoy the effects of prevention of metabolic diseases and prolonging life span by fasting or caloric restriction, as with model animals,” said Teruya. “Understanding the metabolic changes caused by fasting is expected to give us wisdom for maintaining health.”
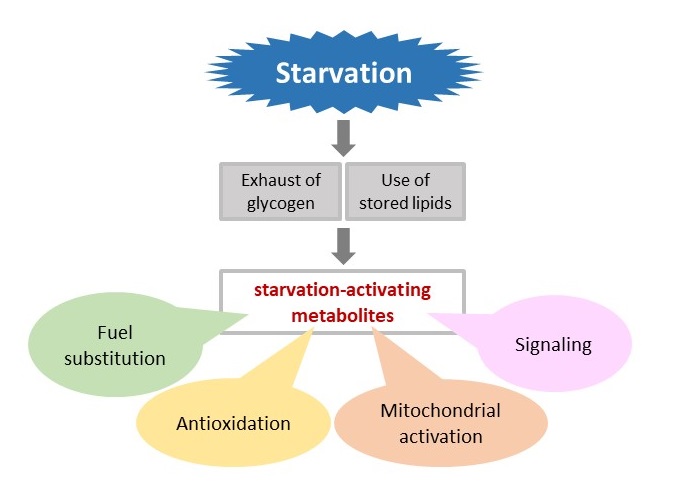
Figure 3. A study by the G0 Cell Unit and Kyoto University researchers suggests that fasting, which puts the body in “starvation mode,” leads to fuel substitution, antioxidation, increased mitochondrial activation and altered signal transduction.
3.4 Casein kinase II-dependent phosphorylation of DNA topoisomerase II suppresses the effect of a catalytic topo II inhibitor, ICRF-193, in fission yeast (From OIST news center article "Anticancer Mechanism Revealed in Yeast Experiments" by Nicoletta Lanese)
Much like shoelaces or dangly necklaces, DNA strands can tangle up in unruly knots. Specialized enzymes keep DNA organized when cells divide, so the cells split smoothly and don’t get stuck. But in tumor cells, this failsafe allows cancer to spread. Now, scientists at the Okinawa Institute of Science and Technology Graduate University (OIST) have learned how this mechanism works in fission yeast and how it might be undermined in human cancer.
The study, published online on January 11, 2019 by the Journal of Biological Chemistry, focuses on a well-established target of cancer therapy: DNA topoisomerase II, or topo II for short. Topo II regulates the topology of DNA, managing the changing structure of the strands over time. Cancerous cells show enhanced topo II activity, which in turn spurs tumor growth. Topo II anticancer drugs suppress the enzyme’s function, prevent tumor cells from replicating and stymie cancer’s spread.
But existing topo II drugs have shown limited success—scientists need to know more about the basic mechanism before they can blunt it effectively.
“Many researchers are working on topo II, but they mainly using human or higher eukaryotic cells,” said Dr. Norihiko Nakazawa, first author of the study and a staff scientist in the OIST G0 Cell Unit, led by Prof. Mitsuhiro Yanagida. The Unit opted to study topo II in fission yeast because its cell division mechanisms are largely conserved in humans, he said, besides being quick and easy to manipulate.
“We can quickly capture the topo II reaction in yeast, then move on to study it in human cells,“ said Nakazawa. “Our work shows that yeast is a good model to screen topo II drugs and would be a quick method to utilize in clinical applications.”
Boosting the Efficacy of Anticancer Drugs
The researchers aimed to learn how topo II operates at a molecular level. At higher magnifications, its effects are obvious; cells with the intact enzyme divide normally, with half their replicated genetic material on each side of the split. Without topo II, the DNA remains tangled in the center of the parent cell, preventing its proper division into two daughter cells.
Prior research suggests that topo II relies on phosphorylation—the addition of a phosphate group at precise locations in its structure. In fission yeast and human cells, the enzyme casein kinase II, also known as CKII, steps in to phosphorylate specific regions of topo II. In this work, Nakazawa identified these regions and found that some remained phosphorylated throughout the entire cell cycle. This constant chemical exchange boosted activity in a region of topo II known as the “ATPase domain.” Though the researchers observed the enhancement, at first, they could not tell how it actually altered topo II function.
The scientists solved the puzzle by throwing a topo II drug into the mix. The drug ICRF-193 works by inhibiting the ATPase activity of topo II, which effectively traps DNA in a tight configuration so it cannot perform enzymatic reactions. But in the presence of CKII, which boosts ATPase activity, the drug is rendered ineffective.
“This [ATPase domain] enhancement broke the anticancer drug effect,” said Nakazawa. “If we block the phosphorylation, this activation doesn’t happen and the drug remains effective.”
Potential Basis for New Therapies
Evidence suggests that topo II and CKII interact in human cells just as they do in fission yeast. Cancerous cells express higher levels of both topo II and CKII than normal tissues, and both enzymes have been associated with abnormal cell proliferation. Now that scientists understand the underlying mechanism in yeast, they can apply that knowledge in future studies in human cancer cells. Someday, the research may lead to new cancer treatments.
“If we inhibit CKII and topo II, it could be a very effective way to inhibit cancer cell division,” said Nakazawa. “This additive effect could be very powerful in cancer therapy.”
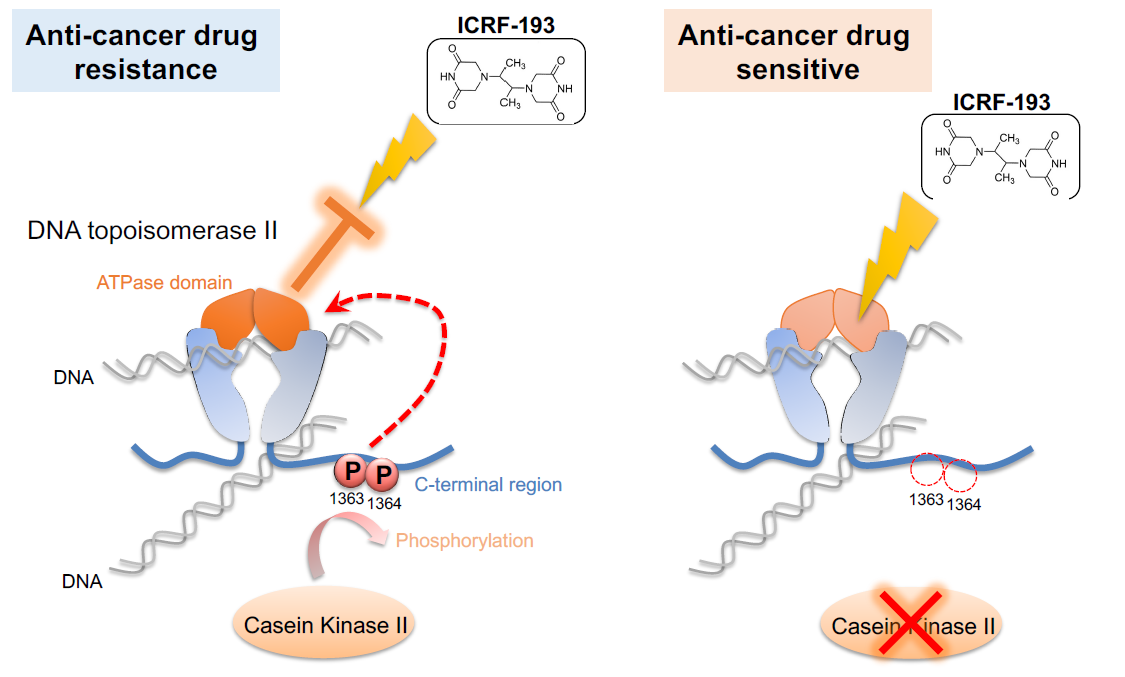
Figure 4. Casein kinase II (CKII) phosphorylates DNA topoisomerase II (topo II), thus activating the ATPase domain of the enzyme (left). This activation cancels the effects of anti-cancer drug ICRF-193. If CKII activity is attenuated (right), the efficacy of ICRF-193 is rescued. The G0 Cell Unit demonstrated this mechanism in fission yeast, a simple model that could be used for anti-cancer drug screening.
4. Publications
4.1 Journals
- Nakazawa N, Arakawa O, Ebe M, Yanagida M (2019) Casein kinase II-dependent phosphorylation of DNA topoisomerase II suppresses the effect of a catalytic topo II inhibitor, ICRF-193, in fission yeast. J Biol Chem [PubMed]
- Teruya T, Chaleckis R, Takada J, Yanagida M, Kondoh H (2019) Diverse metabolic reactions activated during 58-hr fasting are revealed by non-targeted metabolomic analysis of human blood. Scientific Reports 9. Article number 854 [Scientific Reports]
- Sajiki K, Tahara Y, Uehara L, Sasaki T, Pluska T, Yanagida M (2018) Genetic regulation of mitotic competence in G0 quiescent cells. Science Advances. 4 (8): eaat5685 [Science Advances]
- Hayashi T, Teruya T, Chaleckis R, Morigasaki S, Yanagida M (2018) S-Adenosylmethionine Synthetase Is Required for Cell Growth, Maintenance of G0 Phase, and Termination of Quiescence in Fission Yeast. iScience. 5:38-51 [PubMed] [iScience]
- Nakazawa N, Teruya T, Sajiki K, Kumada K, Villar-Briones A, Arakawa O, Takada J, Saitoh S, Yanagida M (2018) The putative ceramide-conjugation protein Cwh43 regulates G0 quiescence, nutrient metabolism and lipid homeostasis in fission yeast. J Cell Sci. [PubMed]
- Suma M, Kitagawa T, Nakase Y, Nakazawa N, Yanagida M, Matsumoto T (2018) Fission Yeast CENP-C (Cnp3) Plays a Role in Restricting the Site of CENP-A Accumulation. G3 (Bethesda). [PubMed]
- Xu X, Kanai R, Nakazawa N, Wang L, Toyoshima C, Yanagida M (2018) Suppressor mutation analysis combined with 3D modeling explains cohesin’s capacity to hold and release DNA. Proc Natl Acad Sci U S A [PubMed]
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yanagida M. Measurement of aging degree using urine and blood by metabolomics analysis and prospect for anti-aging and prevention of diabetes, The 10th Meeting of Anti-Aging Medicine in Urology, Convention Center, Okinawa, 2018.12.09
- Nakazawa N. ER-localized fission yeast protein Cwh43 is essential for both proliferation and G0 quiescence phases, and strongly affects the utilization of principal nutrients as well as lipid metabolism, The 41st Annual Meeting of the Molecular Biology Society of Japan, Pacifico Yokohama, Japan, 2018.11.29
- Nakazawa T, Teruya T, Sajiki K, Xu X, Arakawa O, Saitoh S, Yanagida M. ER-localized fission yeast protein Cwh43 is essential for both proliferation and G0 quiescence phases, and strongly affects the utilization of principal nutrients as well as lipid metabolism, The 41st Annual Meeting of the Molecular Biology Society of Japan, Pacifico Yokohama, Japan, 2018.11.29-11.30
- Sajiki K. CoA metabolism is essential for maintaining mitotic competence under fluctuating nutritional environment, ASBMB Focused Meeting in Coenzyme A in Health, Disease and Bioscience (StellenCoA2018), STIAS, Stellenbosch, South Africa, 2018.10.29
- Yanagida M. CoA in chromatin dynamics, TOR signaling and cwh43 mutant, SASBMB Focused Meeting in Coenzyme A in Health, Disease and Bioscience (StellenCoA2018), STIAS, Stellenbosch, South Africa, 2018.10.29
- Yanagida M. Nuclear and chromosomal proteins required for regulation of G0 phase and proliferation in fission yeast, The 90th Annual Meeting of the Genetic Society of Japan, Nara, Japan, 2018.09.19
- Nakazawa N. Mutations of Mn2+ transporting P-type ATPase, Pmr1, suppress the temperature sensitive mutant of ER-localized ceramide-conjugation protein, Cwh43, in fission yeast, The 51st Annual Meeting of the Yeast Genetics Society of Japan, Kyushu Univ., Fukuoka, Japan, 2018.09.11
- Sajiki K. The role of Nem1 phosphatase at the G0 phase entry, The 51st Annual Meeting of the Yeast Genetics Society of Japan, Kyushu Univ., Fukuoka, Japan, 2018.09.11
- Suma M. Function of fission yeast Mis4 and Mis16 in G0 phase, The 51st Annual Meeting of the Yeast Genetics Society of Japan, Kyushu Univ., Fukuoka, Japan, 2018.09.11
- Suma M. Fission yeast CENP-C(Cnp3) plays a role in restricting the site of CENP-A(Cnp1) accumulation, Gordon Research Conference and Seminar Centromere Biology (GRC), Mount Snow, USA, 2018.07.29-08.03
- Xu X. Negative regulation of Mis6-Mis17 centromere complex by mRNA decay pathway and EKC/KEOPS complex in Schizosaccharomyces pombe, Gordon Research Conference and Seminar Centromere Biology (GRC), Mount Snow, USA, 2018.07.29-08.03
- Yanagida M. Molecular measures of human aging and a new cohesin model, Thermal Biology The 6th Meeting, Okinawa Convension Center, Okinawa, Japan, 2018.06.25
- Yanagida M. Molecular measures of human aging and a new cohesin model, University of Science and Technology of China, Hefei, Anhui, P.R. CHINA, 2018.05.18
- Yanagida M. Molecular measures of human aging and control of cellular quiescence, Shandong University, Shangdong, P.R. CHINA, 2018.05.14
- Yanagida M. Blood metabolite research for human aging, The 91st JES (Japan Endocrine Society) Meeting , Miyazaki, Japan, 2018.04.26
5. Intellectual Property Rights and Other Specific Achievements
- Yanagida M, Teruya T. RISK EVALUTION FOR TYPE 2 DIABETES USING BLOOD METABOLITES AS AN INDEX. Provisional Japanese patent application, JP2018217013 (filing date 20.11.2018)
- Yanagida M, Teruya T. NOVEL TRIMETHYL-AMINO BUTYRATE METABOLITES IN HUMAN BLOOD CELLS THAT INCREASE BY AGING. QUANTITATIVE, SYNTHETIC EVALUATION OF HUMAN AGING BY PCA -PRINCIPAL COMPONENT ANALYSIS- OF AGE-RELATED BLOOD METABOLITES. THE USE OF SALIVA AND URINE METABOLITES FOR HUMAN-AGING BIOMARKERS. Provisional US patent application, US 62/661,666 (filing date 24.04.2018)
6. Meetings and Events
6.1 OIST Seminar
- Title: Cell-cell adhesion machinery conserved across multicellular animals
- Date: October 25, 2010
- Venue: OIST Campus Lab1
- Speaker: Dr. Masatoshi Takeichi, Team Leader, RIKEN Center for Biosystems Dynamics Research
6.2 OIST Pombe Workshop
- Date: February 18-19, 2019
- Venue: OIST Campus Lab1
- Speakers:
- Dr. Benoit Arcangioli, Head of Structure (Dept. of Genomes and Genetics, Institute Pasteur)
- Dr. Karl Ekwall, Professor (Medical Genetics and Epigenetics, Dept. of Biosciences and Nutrition, Karolinska Institutet)
- Dr. Kunihiro Ohta, Professor (Dept. of Life Sciences, The University of Tokyo)
- Dr. Kojiro Ishii, Professor (Kochi Institute University)
- Dr. Junko Kanoh, Associate Professor (Lab. of Nuclear network, Inst. For Protein Research, Osaka University)
- Dr. Mitsuhiro Yanagida, Professor (G0 Cell Unit, OIST)
7. Other
Nothing to report.



