FY2012 Annual Report
Summary
This year marked a transition for our lab, moving from questions centered on just a few organisms, to the development of broadly useful next-generation tools, particularly those aimed at working with degraded DNA and population genetics. In some cases, we can already report success, such as with the recently published study of Okinawan snake venoms (Aird et al, 2013), which used a novel combination of mass spectrometry and next-generation sequencing. Other methodological developments will take a bit of time to bear fruit. However, we are pleased to report the publication of several of our early projects.
New members
Two new members joined our team this year. Misato Okamoto, a post-doctoral fellow, who specializes in ant evolution, and will be working on understanding the genetic control of caste differentiation. Carmen Emborski, who is a 'Special Student' from Texas Tech University. She will do her Ph.D. research work at OIST, focusing on the role of RNA in trans-generational inheritance, and the role of diet and environmental contaminants in obesity.
1. Lab members
- Dr. Alexander Mikheyev, Asistant Professor (PI)
- Dr. Yutaka Watanabe, Researcher
- Dr. Misato Okamoto, Researcher
- Mandy Tin, Technical Staff
- Carmen Emborski, Special Student
- Hitomi Shinzato, Research Administrator
2. Ongoing joint research collaborations (in order of duration)
2.1. Molecular signatures of co-evolution between fungus-gardening
Collaborator
- Ulrich Mueller, University of Texas
2.2. Evolutionary ecology of host-associated differentiation in Ephydryas butterflies
Collaborators
- Michael Singer, University of Texas
- Camille Parmesan, University of Texas, Plymouth University
- Nichole Bennett, University of Texas
2.3. Evolution and composition of snake venoms
Collaborator
- Steven Aird, OIST
2.4. Using museum collections to understand the evolution of feral honeybees in the United States
Collaborator
- Thomas Seeley, Cornell University
2.5. Evolution of social parasitism in harvester ants
Collaborators
- Juergen Gadau, Arizona State University
- Andy Suarez, University of Illinois
- Sara Helms Cahan, University of Vermont
2.6. Genetic control of reproductive division of labor in a queenless Diacamma ants
Collaborator
- Yasukazu Okada, Tokyo University
2.7. Genetic and epigenetic control of termite caste regulation
Collaborators
- Kenji Matsuura, Kyoto University
- Kazuya Kobayashi, Kyoto University
3. Published activities and findings
3.1 Host associated differentiation in Euphydryas butterflies
Many herbivorous, and, in general, parasitic species of insects, are highly host-specific. Host switching, followed by genomic differentiation is a plausible mechanism for the sympatric origin of species. The goal of this series of studies was to understand the interplay between ecological and evolutionary forces that cause the host-associated genetic differentiation (HAD). This research project has been an ongoing collaboration with a team from the University of Texas, focusing on two species of congeneric butterflies, one of which shows a suite of physiological and behavioral adaptations to feeding on a range of host plants (E. editha) and one that does not (E. aurinia). In collaboration with a visiting student, Mel Smee, we used transcriptome sequencing to develop microsatellite markers, for E. aurinia to complement a comparable marker set we previously developed for E. editha (Mikheyev et al. 2010, Smee et al. 2013). Mel was able to implement a rapid and cost-effective way to develop microsatellite markers, completing her project entirely within the span of a two-month rotation. Despite the somewhat unusual method used to prepare these makers, they performed well during cross-validation with another genotyping approach.
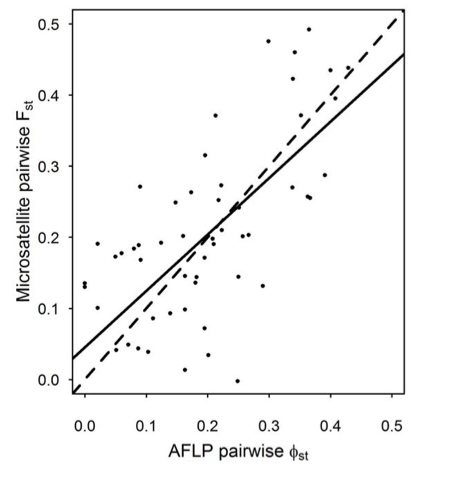
Figure 1. Genetic distances between butterfly populations computed using Mel’s microsatellite markers closely correlated with distances computing using another kind of marker called Amplified Fragment Length Polymorphism (AFLP) markers.
With microsatellite markers in developed for both species, we were able to directly examine the extent of HAD in this system. Although we expected to find the species with the greatest number of host-associated traits to also have the greatest extent of HAD, much to our surprise, we found the exact opposite (Mikheyev et al 2013). Although we were not not able to find a conclusive answer for this finding, this result was generally surprising, and our paper was recommended by the Faculty of 1000. There are a number of hypotheses that could produce such patters, varying from patterns of gene flow, to the strength of selection across the landscape to differences in genomic features of both species, such as the frequency of recombination. We are currently continuing this study, using next-generation molecular markers, in collaborationn with a University of Texas graduate student,
Nichole Bennett, and expect preliminary results in late FY 2013.
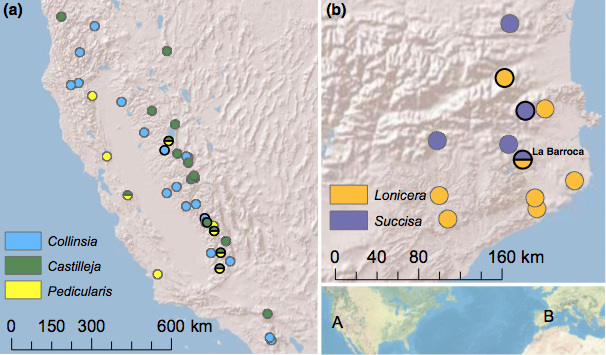
Figure 2. In the Western United States E. editha (a) feeds on at least three host plant species and shows a remarkable suite of host-associated phenotypes (McBride & Singer 2010). In Spain E. aurinia feeds on two host plants, but shows no obvious behavioral or physiological adaptation to either one (Mikheyev et al 2013). However, we could detect no genoimic differentiation in E. editha, whereas strong population differentiation exist in E. aurinia, persisting even in the population that feeds on both hosts, across a period of ten years (population labeled La Barroca).
3.2. Analysis of Okinawan habu snake venom
Together with Steven Aird, who started this study as a professor at University of Maryland University College, and has since migrated to OIST, we wanted to take a high-tech approach to the study of snake venom, focusing on two local species, the habu (Protobothrops flavoviridis) and the himehabu (Ovophis okinavensis). Working with snakes collected by Kouki Terada at the Okinawa Prefectural Institute of Health and Environment, we were able to both extract RNA and venom proteins from the snakes. The RNA and venom was then investigated using RNA sequencing and mass spectrometric analysis. Using an innovative approach developed by Alejandro Villar Birones at the OIST mass spectrometry center, we were able to quantitatively measure protein concentration using mass spec, something that was not possible to do previously (Aird et al, 2013). Even before it was publishedd, this study has attracted attention from Okinawa NHK, which featured a segment on our work, and our own OIST press team.
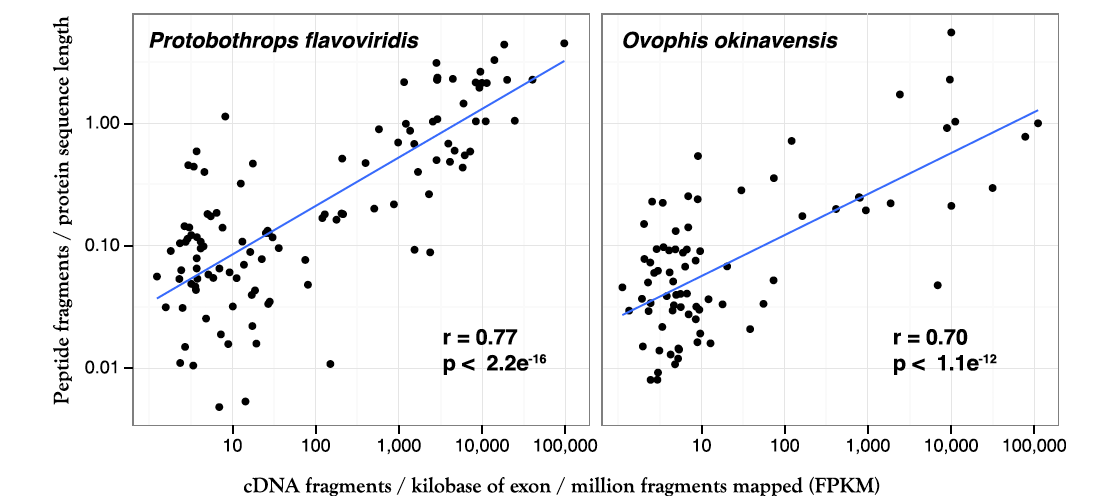
Figure 3. Correlation between RNA sequencing data (x-axis) and mass spectrometric protein data (y-axis) for both species of snakes. The correlation is not perfect, which could be due to several factors both biological, such as different rates of transcription and translation/processing for venom components, and technical, such as differences in sensitivity for the two techniques.
Being able to directly measure and quantitate protein composition in crude venom samples has a wide range of applications. Most importantly, the analysis of RNA requires the extraction of the venom gland, and the sacrifice of the snake, making it impossible to study rare or endangered species. Also, RNA is highly unstable, making it difficult to ship internationally and to conduct comparative studies of snakes in far-flung geographic locations, which is precisely the direction we’re heading next. Our next project, for which we are already collecting venom samples, will be a large comparative study to examine how snakes venom changes with prey type. We also hope to continue our work on local venomous snakes.
4. Publications
4.1. Journals
- Aird, S.D. et al., 2013. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics, 14(1), p.790.
- Mikheyev, A.S. et al., 2013. Host‐associated genomic differentiation in congeneric butterflies: now you see it, now you do not. Molecular Ecology, 22(18), pp.4753–4766.
- Smee, M.R. et al., 2013. Microsatellites for the Marsh Fritillary Butterfly: De Novo Transcriptome Sequencing, and a Comparison with Amplified Fragment Length Polymorphism (AFLP) Markers C. Breuker, ed. PLoS ONE, 8(1), p.e54721.
4.2 Oral and Poster Presentations
- Mikheyev, A.S. “Looking for sex in all the wrong places, and other misadventures in the world of bdelloid rotifers”. Comparative Biology Group, Shanghai, China (2012)
- Mikheyev, A.S. “Transcriptional profiling of attine ant fungal symbionts” British Mycological Society meeting (2012)
- Watanabe, Y. & Mikheyev, A. S. Ongoing sex in bdelloid rotifers, in The 84th Convention of the Genetics Society of Japan, Fukuoka, Japan (2012).
- Watanabe, Y., Brooks, D. & Mikheyev, A. S. “Sequence conservation and gene-conversion; insights into highly * conserved elements in mating-type loci in Saccharomyces cerevisiae”. Society for Molecular Biology and Evolution, Dublin, Ireland (2012).
- Watanabe, Y., Brooks, D. & Mikheyev, A. S. “Sequence conservation and gene-conversion; * insights into highly conserved elements in mating-type loci in Saccharomyces cerevisiae”, 1st Joint Congress on * Evolutionary Biology, Ottawa, Canada (2012).
5. Organized symposia and meetings
5.2 Principles of Insect Fungiculture: Lessons for Human Agriculture?
Date: April 5, 2012
Venue: OIST Campus Lab1
Speaker: Professor Ulrich G. Mueller (The University of Texas at Austin)
5.2. Quantitative Evolutionary and Comparative Genomics (QECG): 2012 Genomic Responses to Selection
Date: August 3–18, 2012
Venue: OIST Seaside House
Organizer: Alexander Mikheyev (OIST)
Co-organizers: Jonathan Miller (OIST) and Patrick Abbot (Vanderbilt University)
Speakers:
- Patrick Abbot (Vanderbilt University)
- David Hillis (University of Texas)
- Jüergen Gadau (Arizona State University)
- Jun Kitano (National Institute of Genetics)
- Lacey Knowles (University of Michigan)
- Michael Lynch (Indiana University)
- Sara Sawyer (University of Texas)
- Kentaro Shimizu (University of Zurich)
- Jay Storz (University of Nebraska)
- Jack Werren (University of Rochester)
- Tetsuji Kakutani (National Institute of Genetics)




