FY2017 Annual Report
Cell Signal Unit
Professor Tadashi Yamamoto

Abstract
The Cell Signal Unit studies various molecular and cellular events that are important for maintaining healthy life in response to environments. The Unit explores the cause of various diseases that include cancer, neuronal disorder, immunological diseases, and diabetes/obesity at the molecular level. To approach these issues, the Unit characterizes regulation of gene expression at the level of RNA that includes micro RNA, mRNA and long non-coding RNA. The Unit also characterizes cell signaling with special interest on control of the brain function such as emotions, learning and memory.
1. Staff
- Dr. Olga Elisseeva, Group Leader
- Dr. Patrick Stoney, Staff Scientist
- Dr. Ken Matsuura, Staff Scientist
- Dr. Akiko Yanagiya, Staff Scientist
- Dr. Guillaume Vares, Staff Scientist
- Dr. Taku Kureha, Postdoctoral Scholar
- Dr. Akinori Takahashi, Postdoctoral Scholar
- Dr. Kristopher Paraiso Montrose, Postdoctoral Scholar
- Dr. William Ashworth, Postdoctoral Scholar
- Dr. Shou Soeda, Postdoctoral Scholar
- Ms. Miho Tokumasu, Technical Staff
- Ms. Tiziana Paganini, Technical Staff
- Ms. Saori Nishijima, Technical Staff
- Ms. Risa Ishida, Technical Staff
- Ms. Melissa Germany, Technical Staff
- Ms. Sandrine Anne Laure Burriel, Graduate Student
- Mr. Haytham Mohamed Aly Mohamed, Graduate Student
- Mr. Shohei Takaoka, Graduate Student
- Mr. Hemanta Sarmah, Graduate Student
- Ms. Dina Mostafa, Graduate Student
- Mr. Mohieldin Magdy Mahmoud Youssef, Graduate Student
- Mr. Hong Huat Hoh, Graduate Student
- Ms. Kaori Yamashiro, Research Unit Administrator (- Nov. 2017)
- Ms. Yuki Nakagawa, Research Unit Administrator (Dec. 2017 -)
2. Collaborations
2.1 Physiological studies of the CCR4-NOT complex
- Description: Analyze the physiological role of each component of the CCR4-NOT complex using gene-modified mice
- Type of collaboration: Joint research
- Researchers:
- Keiji Kuba, Department of Physiology, Graduate School of Medicine, Akita University
2.2 Basic cancer research for prevention and treatment
- Description: Search for substances that contribute to the prevention and treatment of cancer from biological resources, and elucidate its mechanism of action
- Type of collaboration: Joint researches I and II
- Researchers:
- I. Representative director Kuniaki Nerome, Bioresource Laboratory LLC.
- II. Professor Shinya Ikematsu, National Institute of Technology, Okinawa College
2.3 In vivo histology and behavioral analysis of psychiatric model mice
- Description: Analysis of brain structure of 3q29 CNV mice with the state-of-the-art 11.7-Tesla MRI as well as investigating behavior of the mice
- Type of collaboration: Joint research
- Researcher:
- Dr. Kazumasa Yokoyama, Takeda Pharmaceutical Company Ltd.
- Dr. Takanobu Nakazawa, School of Pharmaceutical Science, Osaka University
2.4 Electrophysiology
- Description: Electrophysiological studies of gene modified mice with abnormal social behavior
- Type of collaboration: Joint research
- Researcher:
- Dr. Toshiya Manabe, Institute of Medical Science, University of Tokyo
2.5 Mechanisms and physiological roles of CCR4-NOT complex-regulated gene expression in the brain
- Description: Investigation of mechanisms and physiological roles of CCR4-NOT complex-regulated gene expression in the brain
- Type of collaboration: Joint research
- Researcher:
- Team leader Masaru Tamura, RIKEN
2.6 Study of Cell-to-cell Interaction within Tumor Microenvironment Using An In-Vitro Three Dimensional Pancreatic Cancer Model
- Description:
- Establishing an in vitro 3D organoid culture for pancreatic ductal adenocarcinoma to study cell-cell interaction within tumor microenvironment
- Investigating the origin of cancer stem cells in pancreatic ductal adenocarcinoma by live cell imaging using tumor organoid
- Characterizing the effects of the interaction between stromal cells and cancer stem cells
- Identifying the targets for tumor-stroma cross-talk and cancer stem cells
- Type of collaboration: Joint research
- Researchers:
- Professor Masafumi Nakamura, Assistant Professors Kenoki Ohuchida, Kazuhiro Koikawa, Department of Surgery and Oncology, Graduate School of Medical Sciences, Kyushu Universit
2.7 Analysis of CNOT9 function during mouse development by micro-CT imaging
- Description: Detailed analysis of embryonic defects in CNOT9 KO mice using micro CT imaging
- Type of collaboration: Joint research
- Researcher:
- Team leader Masaru Tamura, RIKEN
3. Activities and Findings
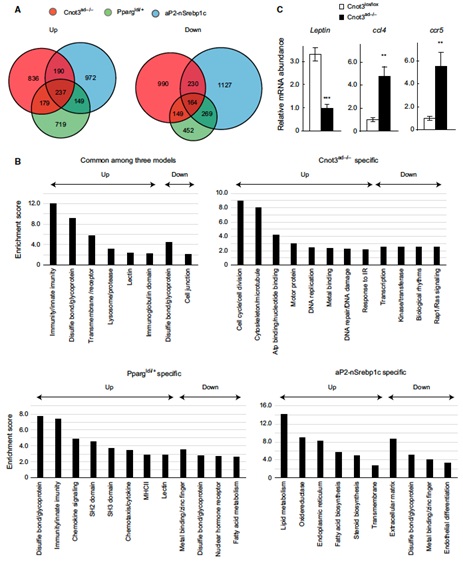
3.1 Adipocyte-specific disruption of mouse Cnot3 causes lipodystrophy.
Lipodystrophy involves a loss of adipose tissue. In mice, disruption of adipose tissue Cnot3, a subunit of the CCR4-NOT deadenylase complex, causes adipose tissue anomalies. In Cnot3ad-/- mice, white adipose tissue (WAT) decreases concomitantly with enhanced inflammation, whereas brown adipose tissue increases and contains larger lipid droplets. Cnot3ad-/- mice show hyperinsulinemia, hyperglycemia, insulin resistance, and glucose intolerance, and cannot maintain body temperature during cold exposure. Increased expression of inflammatory genes and decreased leptin expression also occur in Cnot3ad-/- WAT, achieving levels similar to those in lipodystrophic aP2-nSrebp1c and Ppargldi/+ mice; thus, Cnot3ad-/- mice exhibit lipodystrophy.
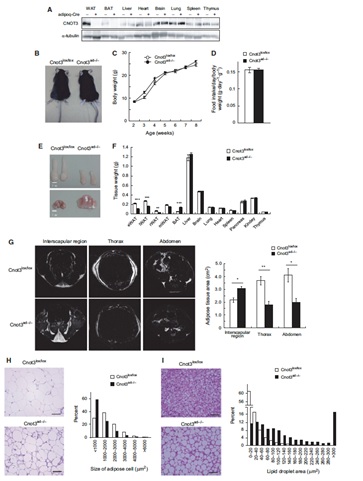

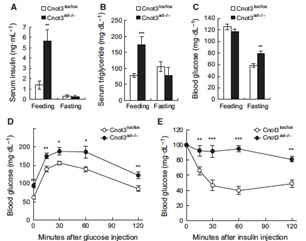

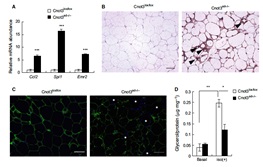

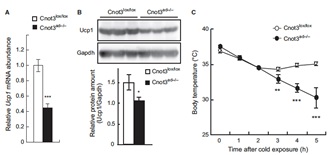
![]()

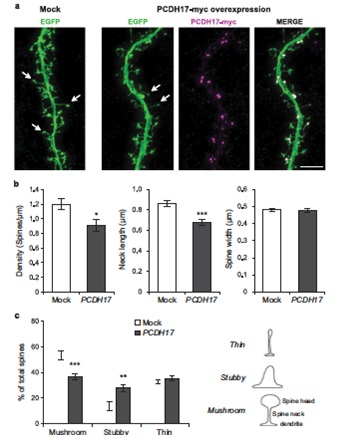
3.2 The protocadherin 17 gene affects cognition, personality, amygdala structure and function, synapse development and risk of major mood disorder
Major mood disorders, which primarily include bipolar disorder and major depressive disorder, are the leading cause of disability worldwide and pose a major challenge in identifying robust risk genes. Here, we present data from independent large-scale clinical data sets (including 29 557 cases and 32 056 controls) revealing brain expressed protocadherin 17 (PCDH17) as a susceptibility gene for major mood disorders. Single-nucleotide polymorphisms (SNPs) spanning the PCDH17 region are significantly associated with major mood disorders; subjects carrying the risk allele showed impaired cognitive abilities, increased vulnerable personality features, decreased amygdala volume and altered amygdala function as compared with non-carriers. The risk allele predicted higher transcriptional levels of PCDH17 mRNA in postmortem brain samples, which is consistent with increased gene expression in patients with bipolar disorder compared with healthy subjects. Further, overexpression of PCDH17 in primary cortical neurons revealed significantly decreased spine density and abnormal dendritic morphology compared with control groups, which again is consistent with the clinical observations of reduced numbers of dendritic spines in the brains of patients with major mood disorders. Given that synaptic spines are dynamic structures which regulate neuronal plasticity and have crucial roles in myriad brain functions, this study reveals a potential underlying biological mechanism of a novel risk gene for major mood disorders involved in synaptic function and related intermediate phenotypes.
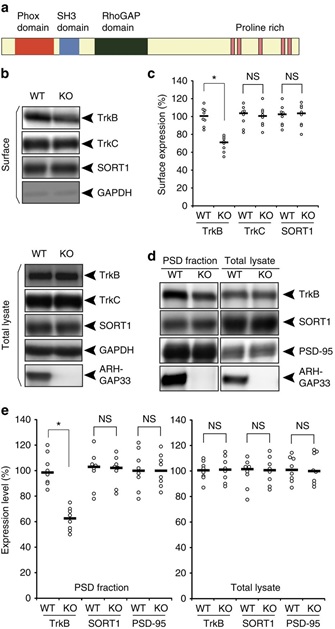
(a) Protein structure of a brain-enriched SNX protein, ARHGAP33. ARHGAP33 has an N-terminal PX domain, an SH3 domain and a RhoGAP domain. (b,c) Decreased cell-surface expression of TrkB in ARHGAP33 KO mice. Biotinylated cell-surface proteins (upper) and total lysates (lower) of WT and ARHGAP33 KO neurons (14 DIV) were immunoblotted with anti-TrkB, anti-TrkC, anti-SORT1, anti-GAPDH and anti-ARHGAP33 antibodies. (b) Representative blots. (c) Quantification of surface expression (each, n=8; TrkB, P=7.8 × 10−4; TrkC and SORT1, P>0.05; Mann–Whitney U-test). The expression levels of TrkB, TrkC and SORT1 in ARHGAP33 KO neurons were normalized to those in WT neurons (The averaged WT values were set to 100%). (d,e) Decreased TrkB in the isolated PSD fraction of ARHGAP33 KO mice. The isolated PSD fraction and total lysates of WT and ARHGAP33 KO mice were immunoblotted with anti-TrkB, anti-PSD-95, anti-SORT1, and anti-ARHGAP33 antibodies. Representative blots (d). Quantification for the isolated PSD fraction (each, n=8, TrkB, P=7.7 × 10−4; SORT1 and PSD-95, P>0.05; Mann–Whitney U-test) and for the total lysate (each, n=8, P>0.05; Mann–Whitney U-test; e) The expression levels of TrkB, SORT1 and PSD-95 in the PSD fraction and total lysate from ARHGAP33 KO mice were normalized to those from WT mice (The averaged WT values were set to 100%). Note that the amounts of PSD-95 and SORT1 in the isolated PSD fraction from ARHGAP33 KO mice were not significantly different from those in the fraction from WT mice. *P<0.05. NS, not significant. Bars show median values. All western blots show representative results from eight independent experiments performed using different mice.
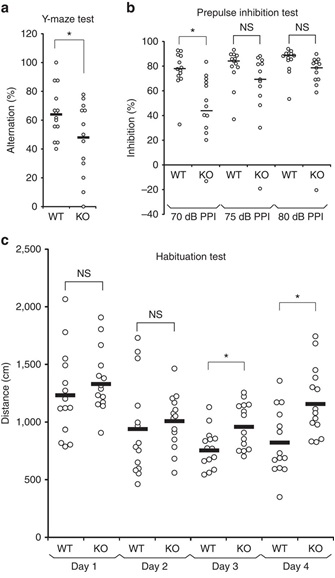
(a) Impaired spontaneous alternations of ARHGAP33 KO mice during the Y-maze test (WT, n=14, KO, n=13, F1,25=5.18, P=0.031, one-way ANOVA). *P<0.05. Bars show mean values. (b) Impaired PPI of ARHGAP33 KO mice (each n=13, P=0.046, Friedman test followed by Scheffe tests). *P<0.05. NS, not significant. Bars show median values. (c) Impaired open field habituation of ARHGAP33 KO mice during the openfield habituation test (each n=14, genotype effect, F1,26=5.08, P=0.033, two-way ANOVA with repeated measures; day 3, P=1.3 × 10−4, day 4, P=7.0 × 10−4, Tukey–Kramer post hoc tests). *P<0.05. NS, not significant. Bars show mean values.
4. Publications
4.1 Journals
-
Chang H, Hoshina N, Zhang C, Ma Y, Cao H, Wang Y, Wu D, Bergen E, Landén M, Hultman C, Preisig M, Kutalik Z, Castelao E, Grigoroiu-Serbanescu M, Forstner A, Strohmaier J, Hecker J, Schulze T, Müller-Myhsok B, Reif A, Mitchell P, Martin N, Cichon S, Nöthen M, The Swedish Bipolar Study Group, MooDS Bipolar Consortium, Walter H, Erk S, Heinz A, Meyer-Lindenberg A, Tost H, Xiao X, Yamamoto T, Rietschel M and Li M. The Protocadherin 17 Gene Affects Cognition, Personality, Amygdala Structure and Function, Synapse Development and Risk of Major Mood Disorders. Mol Psychiatry 23:400-412, 2018
-
Yamaguchi T, Suzuki T, Sato T, Takahashi A, Watanabe H, Kadowaki A, Miyuki N, Inagaki H, Arakawa S, Nakaoka S, Koizumi Y, Seki S, Adachi S, Fukao A, Fujiwara T, Natsume T, Kimura A, Komatsu M, Shimizu S, Ito H, Suzuki Y, Penninger JM, Yamamoto T, Imai Y and Kuba K. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Science Signalinig 11: eaan3638, 2018
-
Li T, Amari T, Semba K, Yamamoto T and Takeoka S. Construction and evaluation of pH-sensitive immunoliposomes for enhanced delivery of anticancer drug to ErbB2 over-expressing breast cancer cells. Nanomedicine 13(3):1219-1227,2017
-
Li X, Morita M, Kikuguchi C, Takahashi A, Suzuki T and Yamamoto T. Adipocyte-specific disruption of mouse Cnot3 causes lipodystrophy. FEBS Letters 591(2):358-368,2017
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
-
Huat Hoh, H., Kureha, T, Yamamoto, T. Downregulation of HB-EGF by microRNA-96 leads to cell cycle arrest and reduces cancer stem cells (CSCs) in breast cancer cells. Consortium of Biological Sciences 2017, Kobe, Japan, 2017.12.09 (2017)
-
Yanagiya, A, Albert, O, Robaire, B, Sonenberg, N, Yamamoto, T. Post-transcriptional regulation via the mRNA poly(A) tail during spermatogenesis. Consortium of Biological Sciences 2017, Kobe, Japan, 2017.12.08 (2017)
-
Soeda, S, Ohsugi, M. Molecular mechanism and physiological importance of the temporal regulation of pronuclear formation onset in mouse zygotes. Consortium of Biological Sciences 2017, Kobe, Japan, 2017.12.08 (2017)
-
Montrose, K, Kobayashi, S, Yamamoto, T. Lmtk3 knockout mice exhibit schizophrenic-like behaviour due to impaired GluA1 trafficking. Consortium of Biological Sciences 2017, Kobe, Hyogo, Japan, 2017.12.07 (2017)
-
Stoney, P, Fang, K, Yamamoto, T. A role for the CCR4-NOT deadenylase complex in the brain. Consortium of Biological Sciences 2017, Kobe, Japan, 2017.12.07 (2017)
-
Takahashi, A, Haytham Mohamed M.A., Yamamoto, T. mRNA degradation induces obese gene expression. Consortium of Biological Sciences 2017, Kobe, Japan, 2017.12.07 (2017)
-
Sarmah, H, Ito, K, Kikuguchi, C, Yamamoto, T. Molecular and Physiological Function of Mammalian CNOT9. Cutting Edge Developments in RNA Biology for the Control of Gene Expression, OIST, Japan, 2017.11.17 (2017)
-
Yamamoto, T. Introductory overview: RNA biology 2017. Cutting Edge Developments in RNA Biology for the Control of Gene Expression, OIST, Japan, 2017.11.13 (2017)
-
Yamamoto, T. Post-transcriptional regulation by mRNA poly(A) tail. U of RYUKYUS & OIST JOINT SYMPOSIUM 2017, OIST, Japan, 2017.10.31 (2017)
-
Takahashi, A, Yamamoto, T. Importance of mRNA decay in the regulation of obesity. The 38th Annual Meeting of Japan Society for the Study of Obesity, 2017.10.7 (2017)
-
Huat Hoh, H, Vares, G, Yamamoto, T. Mesenchymal stem cells reprogram pancreatic cancer into cancer stem cell phenotype via direct contact interaction. The 76th Annual Meeting of the Japan Cancer Association, Yokohama, Japan, 2017.09.29 (2017)
-
Tokumasu M, Kureha T, Yamamoto M, Nakayama J, Semba K, Inoue J, Yamamoto T. Negative regulation of NF-kB signaling by Tob1 in breast cancer. The 76th Annual Meeting of the Japanese Cancer Association, 2017.9.28 (2017)
-
Takahashi, A. Elucidation of the regulation of obesity through mRNA degradation. The 10th Symphony, MSD, Tokyo, Japan, 2017.09.17 (2017)
-
Takaoka S, Takahashi A, Mohamad, Hytham, M. A, Ashworth, W, Asai Y, Yamamoto T. Mathematical analysis of the regulation of circadian rhythm through mRNA degradation. The 1st Workshop for Young Researchers: Integrative understanding of biological signaling networks based on mathematical science for Grant-in-Aid for Scientific Research on Innovative Areas, Shizuoka, Japan, 2017.08.07 (2017)
-
Mohamed, Haytham, M.A, Takahashi, A, Yamamoto, T. CNOT1 regulation of eukaryotic circadian clock through post-transcriptional mechanisms. 19th Annual Meeting of the RNA Society of Japan , Toyama, Japan, 2017.07.21 (2017)
-
Yanagiya, A, Albert, O, Downey, A. M, Robaire, B, Sonenberg, N. Translational control in spermatogenesis by poly(A)-binding protein-interacting protein 1. The 19th Annual Meeting of the RNA Society of Japan, RNA meeting 2017, Toyama, Japan, 2017.07.21 (2017)
-
Takahashi, A, Takaoka S, Yamaguchi T, Kuba K, Yamamoto T. Deadenylation regulates mRNA decay rate and transcription activity. The RNA Society of Japan 2017, Toyama, Japan, 2017.07.20 (2017)
-
Takahashi, A. Mathematical analysis of the circadian rhythm regulation by mRNA degradation. Conference for Integrative understanding of biological signaling networks based on mathematical science for Grant-in-Aid for Scientific Research on Innovative Areas, The Institute of Medical Science, The University of Tokyo, Japan, 2017.06.20 (2017)
-
Takahashi, A. Deadenylation regulates the balance between mRNA stability and transcription activity. The 9th Signal Network Study Meeting, RIKEN, Yokohama, Japan, 2017.06.10 (2017)
-
Yamamoto, T. The CCR4-NOT deadenylase: its roles in controlling cells survival and differentiation in various biological systems. RIKEN–Max Planck Joint Research Center for Systems Chemical Biology The 6th Symposium, Pacific Hotel Okinawa, Japan, 2017.4.25 (2017)
5. Intellectual Property Rights and Other Specific Achievements
5.1 Intellectual Property RightsTitle: Anti-tumor agent
Patent Number: JP6443872B2
Inventors: Kuniaki Nerome, Tadashi Yamamoto, Taku Kureha, Reiko Nerome
Owner: The Institute of Biological Resources
Priority Date: Nov. 10, 2016
6. Meetings and Events
6.1 Seminar
6.1.1 Motor circuits for locomotor and skilled movements
- Date: June 5, 2017
- Venue: OIST Campus Lab1
- Speaker: Dr. Yutaka Yoshida (Cincinnati Children’s Hospital Medical Center)
6.1.2 Systems proteomics of liver mitochondrial activity
- Date: August 22, 2017
- Venue: OIST Campus Lab1
- Speaker: Dr. Yibo Wu (RIKEN)
6.1.3 HLA, Immune Response and Disease - Classical and New Concept -
- Date: November 7, 2017
- Venue: OIST Campus Lab1
- Speaker: Dr. Takehiko Sasazuki (Kyusyu University)
6.1.4 Molecular grounds underlying chromosomal instability in cancers
- Date: January 13, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Toru Hirota (Japanese Foundation for Cancer Research)
6.1.5 PolyADP-ribosylation and cell proliferation
- Date: February 7, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Masanao Miwa (Nagahama Bio University)
6.1.6 The TRAF6-NF-kB-NFATc1 signal pathway in RANK-induced osteoclastogenesis
- Date: February 14, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Jun-ichiro Inoue (Institute of Medical Science, The University of Tokyo)
6.1.7 Genetic mechanism involving PD ligands to promote immune evasion in multiple cancers
- Date: February 28, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Keisuke Kataoka (National Cancer Center Research Institute)
6.1.8 Identification of small molecule inhibitors of stromal fibroblast cell migration accelerated by cancer cells
- Date: March 7, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Nobumoto Watanabe (RIKEN)
6.1.9 IER5 is a p53-reguated activator of HSF1 that contributes to promotion of cancer
- Date: March 14, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Rieko Ohki (National Cancer Center Research Institute)
6.1.10 Cooperative function of genes for cellular transformation and an evaluation system of tumorigenic activity
- Date: March 28, 2018
- Venue: OIST Campus Lab1
- Speaker: Dr. Kentaro Semba (Waseda University)
6.2 Meeting
6.2.1 OIST-JST Joint Seminar
- Date: November 28, 2017
- Venue: OIST Meeting Rooms
- Co-organizers: Japan Science Technology (JST)
- Speakers:
- Dr. Nicholas M. Luscombe (OIST)
- Dr. Kenji Doya (OIST)
- Dr. Yoko Yazaki-Sugiyama (OIST)
- Dr. Ryoichiro Kageyama (Kyoto University)
- Dr. Shigeru Kondo (Osaka University)
6.2.2 RNA Biology 2017
- Date: November 13-17, 2017
- Venue: OIST Meeting Rooms
- Speakers:
- Dr. Toshinobu Fujiwara (Kindai University)
- Dr. Matthias Hentze (EMBL)
- Dr. Osamu Takeuchi (Kyoto University)
- Dr. Shintaro Iwasaki (RIKEN)
- Dr. Toshifumi Inada (Tohoku University)
- Dr. Thomas Preiss (Australian National University)
- Dr. Clement Chapat (McGill University)
- Dr. Martine Collart (University of Geneva)
- Dr. Keiji Kuba (Akita University)
- Dr. Sebastiaan Winkler (The University of Nottingham)
- Dr. David Gatfield (University of Lausanne)
- Dr. Yukako Chiba (Hokkaido University)
- Dr. Hidetoshi Saze (OIST)
- Dr. Yukihide Tomari (The University of Tokyo)
- Dr. Joseph Reese (Penn State University)
- Dr. Mark Bartlam (Nankai University)
- Dr. Bertrand Seraphin (Institute of Genetics and Molecular and Cellular Biology (IGBMC))
- Dr. Teruko Tamura-Nieman (Hannover Medical School)
- Dr. Yumiko Imai (NIBIOHN)
- Dr. Masatoshi Hagiwara (Graduate School of Medicine, Kyoto University)
- Dr. Kent Duncan (Center for Molecular Neurobiology Hamburg)
- Dr. Charles Joel McManus (Carnegie Mellon University)
- Dr. Yuichiro Mishima (The University of Tokyo)
- Dr. Charles Plessy (RIKEN)
7. Other
Nothing to report.



