FY2012 Annual Report
Cell Signal Unit
Professor Tadashi Yamamoto

Abstract
The Cell Signal Unit studies various molecular and cellular events that are important for maintaining healthy life in response to environments. The Unit explores the cause of various diseases that include cancer, neuronal disorder, immunological diseases, diabetes/obesity at the molecular level. More specifically, the Unit characterizes molecular biologically a set of proteins and microRNAs that regulate and execute degradation of mRNAs to silence gene expression. The Unit also studies the mechanisms by which protein kinases and cell adhesion molecules control the brain function such as emotions, learning and memory.
1. Staff
- Dr.Toru Suzuki, Group leader
- Dr.Naosuke Hoshina, Researcher
- Dr.Yo-taro Shirai, Researcher
- Dr.Taku Kureha, Researcher
- Dr.Akinori Takahashi, Researcher
- Ms.Miyuki Hoshina, Technical Staff
- Ms.Chisato Kikuguchi, Technical Staff
- Ms.Xue Li, Technical Staff
- Ms.Kanako Yamauchi, Staff
- Ms.Kaori Yamashiro, Research Administrator
2. Collaborations
- Theme: Physiological studies of the CCR4-NOT complex
- Type of collaboration: Joint research
- Researchers:
- Kuba K and Imai Y. Department of Physiology, Graduate School of Medicine, Akita University
- Theme: Structural analysis of the CCR4-NOT complex
- Type of collaboration:Joint research
- Researchers:
- Bartlam M and Rao Z. College of Life Sciences, Nankai University, China
- Theme: Molecular mechanism of mRNA degradation mediated by the CCR4-NOT deadenylase
- Type of collaboration: Joint research
- Researchers:
- Morita M, Fabian M, and Sonenberg N. Department of Biochemistry and Goodman Cancer Research Center, McGill University, Canada
- Theme: Bioinformatics of gene expression affected by impairment of mRNA degradation machinery
- Type of collaboration: Joint research
- Researchers:
- Nagashima T and Okada M. Laboratory for Cellular System Modeling, RIKEN Research Center for Allergy and Immunology
- Theme: Roles of the Fyn tyrosine kinase and its substrates in the central nervous system
- Type of collaboration: Joint research
- Researchers:
- Nakazawa T and Kano M. Department of Neurophysiology, Graduate School of Medicine, University of Tokyo
-
Theme: Roles of the CNOT3 subuni of the CCR4-NOT complex in B cell development
- Type of collaboration: Joint research
- Researchers:
- Inoue T and Kurosaki T. Immunology Frontier Research Center, Osaka University
3. Activities and Findings
Our current interest is to characterize cell signaling that are relevant to physiological properties of mammals such as cancer development, diabetes, immunity and neuronal function.
1. The biological role of Tob family proteins and CCR4-NOT complex.
(i) Studies on the Tob family of proteins.
In the study to dissect signaling pathway downstream of ErbB/EGF receptor family tyrosine kinases, we identified the tob family of genes that code for proteins, Tob, Tob2, and ANA, that are associated with anti-proliferative function. Upon EGF stimulation MAP kinase became activated downstream of the EGF receptor. We found that MAP kinase in turn phosphorylated Tob, resulting in suppression of the antiproliferative activity of Tob. Further analyses of Tob family proteins with gene manipulated mouse lines revealed followings. #1: Mice lacking Tob were prone to develop cancer. #2: Mice lacking Tob showed osteopetrosis-like phenotype due to enhanced differentiation and proliferation of osteoblasts. In contrast, #3, mice lacking Tob2 had decreased bone mass due to the increment of osteoclasts. #4: Tob2-deficinet mice exhibit increased adiposity with augmented mass of the epididymal WAT. #5: Mice lacking ana, which was specifically expressed in type II alveolar epithelial cells, developed spontaneous lung adenocarcinoma. The underlying molecular mechanisms of these phenotypes have been addressed to reveal that Tob negatively regulates cyclin D expression. Tob also negatively regulate BMP (bone morphogenetic protein)-mediated signaling. Tob2 negatively regulate formation of osteoclasts by suppressing RANKL expression through its interaction with Vitamin D3 receptor. In addition, Tob2 could negatively regulate adipogenesis by inhibiting PPARg2 expression (Fig. 1).
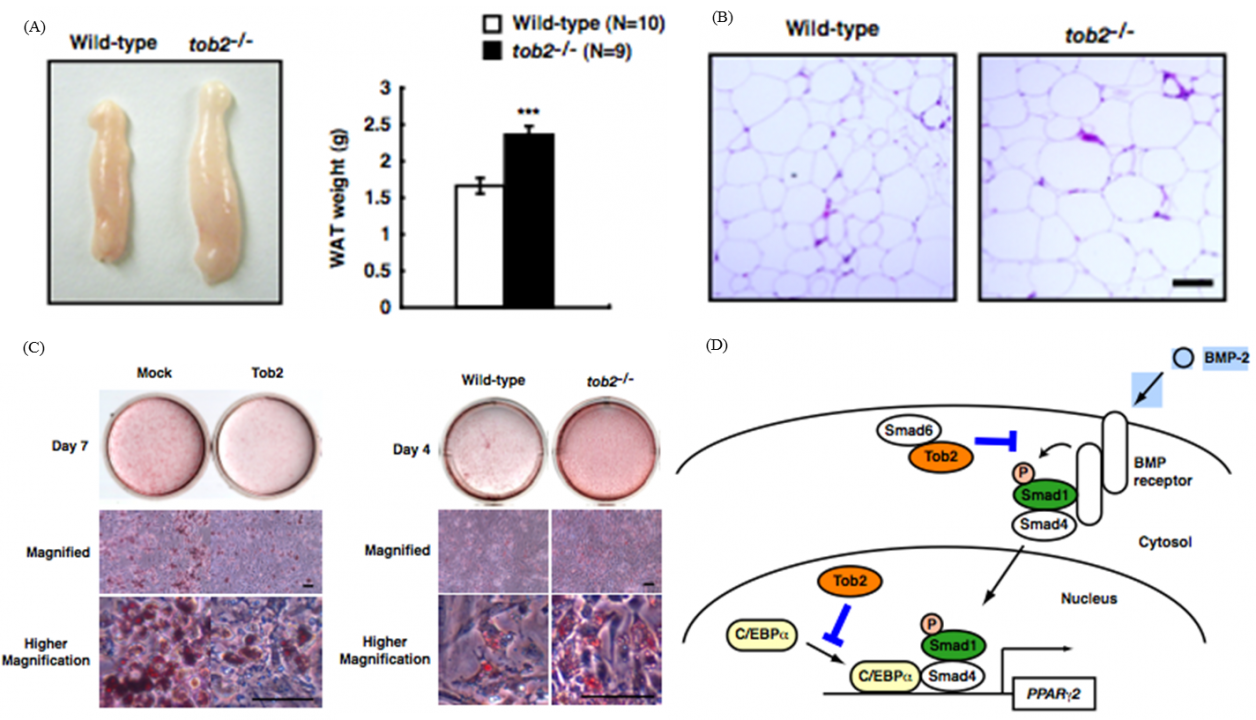
Fig. 1 Tob2 negatively regulates adipogenesis and BMP-2 dependent PPARg2 induction
(A) Macroscopic view (left) and weight (right) of the WT and tob2-/- mice. (B) H&E staining of the epididymal WAT in WT and tob2-/- mice.(C) Oil Red O staining of WAT-derived preadipocytes. (left) Comparison between Mock transfected cells and Tob2-overexpressed cells. (right)Comparison between the cells prepared from WR and tob2-/- mice. (D) Model for the role of Tob2 in BMP-2 signaling.
Apart from these anti-proliferative and/or anti-differentiation characters, Tob exhibits anti-apoptotic properties in response to DNA damage. UV (high dose)-induced stress promotes proteasome-dependent degradation of Tob, triggering an apoptotic signal. Low-doses of UV do not trigger apoptosis and do not promote Tob degradation either. Rather, Low-doses of UV stabilize Tob. Suppression of Tob by small-interfering RNAs results in frequent induction of apoptosis in response to low doses of UV. This induction of apoptosis occurs irrespectively to the presence or absence of functional p53. Taken together, clearance of Tob provides a novel p53-independent pathway for UV-induced apoptosis. Involvement of proteasomes in degradation of Tob has been reported. We found that Cul4-DDB1(Cdt2) induces ubiquitination and degradation of Tob. Interestingly, DNA replication-initiating kinase Cdc7 inhibits Cul4-DDB1(Cdt2)-induced Tob degradation (Fig. 2). Cdc7-mediated Tob phosphorylation appears to contribute to suppression of a low dose of UV-induced apoptosis.
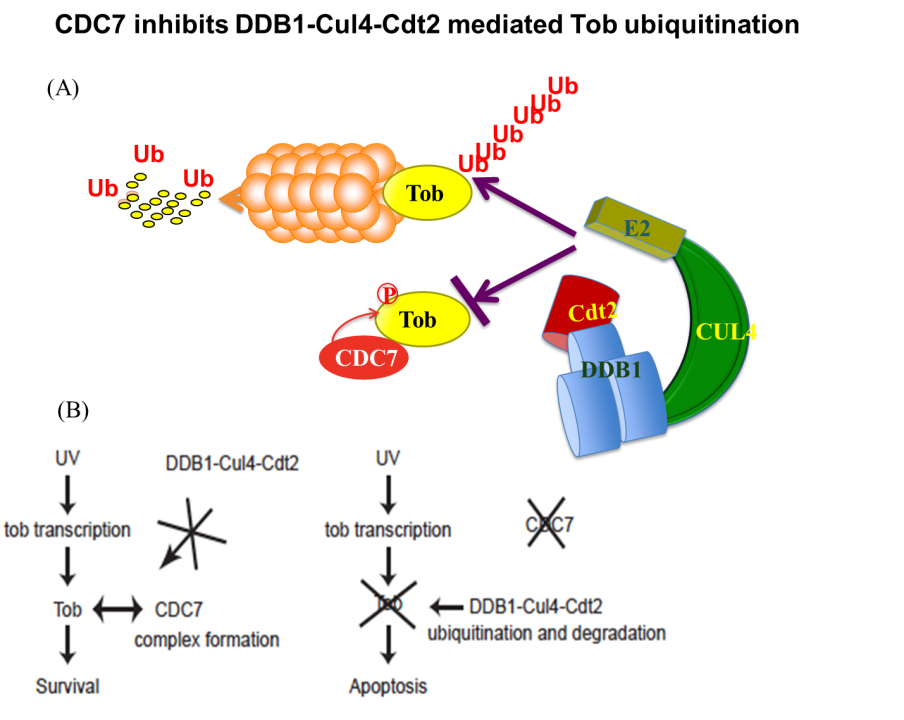
Fig. 2 (A) CDC7 protects Tob from ubiquitination–dependent proteasomal degradation. (B) Tob is inhibitory against UV-induced apoptosis.
(ii) Studies on the CCR4-NOT deadenylase complex.
Apparently, Tob exhibits multiple functions. To address the molecular mechanisms that would explain the multi-functionality, we carried out mass spectrometric analysis of the Tob-interacting proteins prepared from the lysates of HeLa cells and identified CNOT proteins that formed a large (2.0 MDa and 1.2 MDa) complex called CCR4-NOT. The CCR4-NOT complex is conserved from yeast to human and is associated with the mRNA deadenylase activity. In yeast, two components of the complex, Ccr4p and Caf1p, possesses the deadenylase activity. The mammalian orthologs of Ccr4p are CNOT6 and CNOT6L, and those of Caf1p are CNOT7 and CNOT8. Our recent structural analysis of the CNOT6L complexed with nucleotides revealed a deadenylation mechanism involving a pentacovalent phosphate transition. As Tob also interacts with poly(A)-binding protein (PABP), we assume that Tob could be involved in recruitment of the CCR4-NOT complex to the poly(A) sequence at the 3’end of mRNAs (Fig. 3).
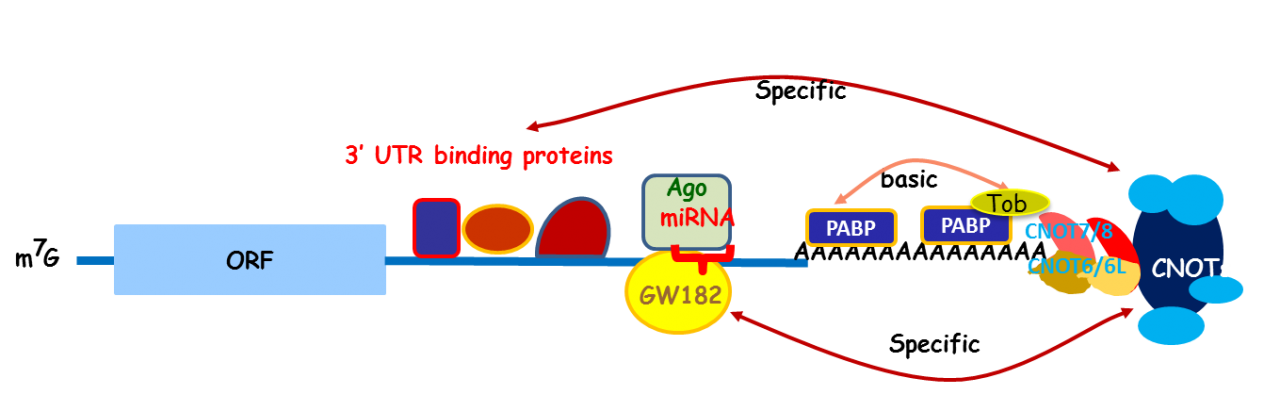
Fig. 3 A role of Tob in recruitment of the CCR4-NOT deadenylase to mRNA poly(A) tails
To help understand the physiological significance of the CCR4-NOT deadenylase complex in mammals, we have knocked-down the expression of each subunit by treating the cultured cells with shRNA and/or siRNA. Depletion of CNOT1 or CNOT2 induced protein overexpression due to the stabilization of many mRNA species, resulting in the ER stress-induced apoptotic death. Depletion of CNOT6L from NIH3T3 cells specifically augmented the stability of p27kip1 mRNA, causing cell cycle arrest at the G1 phase. CNOT3 depletion in cultured cells induced elevation of the mad1 mRNA level and impaired the M-phase checkpoint response. It remains to be elucidated how the CCR4-NOT deadenylase complex recognizes the specific mRNAs.
To examine the in vivo functions of CCR4-NOT deadenylase complex, we generated gene-engineered mouse lines each lacking individual cnot genes. We found that deficiency of every cnot gene, except cnot6, cnot6L and cnot7, causes embryonic death at distinct developmental stages. Mice lacking cnot1, 2, 3, 8, 9 and 10, respectively die at embryonic day around 7.5 (E7.5), E15.5, E6.5, E9.5, and E10.5. The cause of death appears to be different between the mice lines. For example, cnot3-/- mice die due to the defect of early embryogenesis and cnot9-/- mice due to poor vasculogenesis. Though cnot7-/- mice appears normal, male are sterile due to poor spermatogenesis. Phenotypes of cnot6-/- and cnot6L-/- mice are under extensive analysis. The results tentatively suggest that gene expression is distinctively affected in each mouse lines and that non-enzymatic subunits of the CCR4-NOT complex are relevant to regulate target specificity of the CCR4-NOT complex.
Although cnot3-/- mice are embryonic lethal, cnot3+/- mice are lean with hepatic and adipose tissues containing reduced levels of lipids, and show increased metabolic rates and enhanced glucose tolerance. cnot3+/- mice remain lean and sensitive to insulin even on a high-fat diet. Hepatic expression of most mRNAs is not altered in cnot3+/- vis-à-vis wild-type mice. However, the levels of specific mRNAs, such as those coding for energy metabolism-related PDK4 and IGFBP1, are increased in cnot3+/- hepatocytes, having poly(A) tails that are longer than those seen in control cells. We provided evidence that CNOT3 is involved in recruitment of the CCR4-NOT deadenylase to the 3' end of specific mRNAs. Similarly, change of expression of various mRNAs in mice lacking other subunits has been monitored by employing expression microarray analysis, Real-time PCR, and Northern blot analysis. A subset (not all) of mRNA species was increased in each gene-engineered mouse, which is consistent with the idea that the CNOT complex targets specific sets of mRNAs for deadenylation depending on temporal and special conditions. Efforts to elucidate the molecular mechanism by which the CCR4-NOT complex recognizes specific mRNAs are ongoing (Fig. 4).
To facilitate efficient conduction of the research, we are currently producing conditional knock-out mouse lines by using mice carrying the cre gene under the control of various transcriptional promoters. So far, we have generated mouse lines in which cnot1, cnot3, and cnot8 are individually depleted specifically in the liver (liver-specific KO of cnot1, cnot3, and cnot8, respectively), in the lung (lung-specific KO of cnot3), in the white adipose tissue (WAT-specific KO of cnot3), in the lymphocytes (B cell-specific and T cell specific KOs of cnot3), and in the brain (brain-specific KO of cnot3). All these mouse lines exhibit interesting phenotypes that are under extensive analysis. Preliminarily, we noticed that expression of possible CCR4-NOT target mRNAs is up-regulated in these conditional knock-out mice lines.
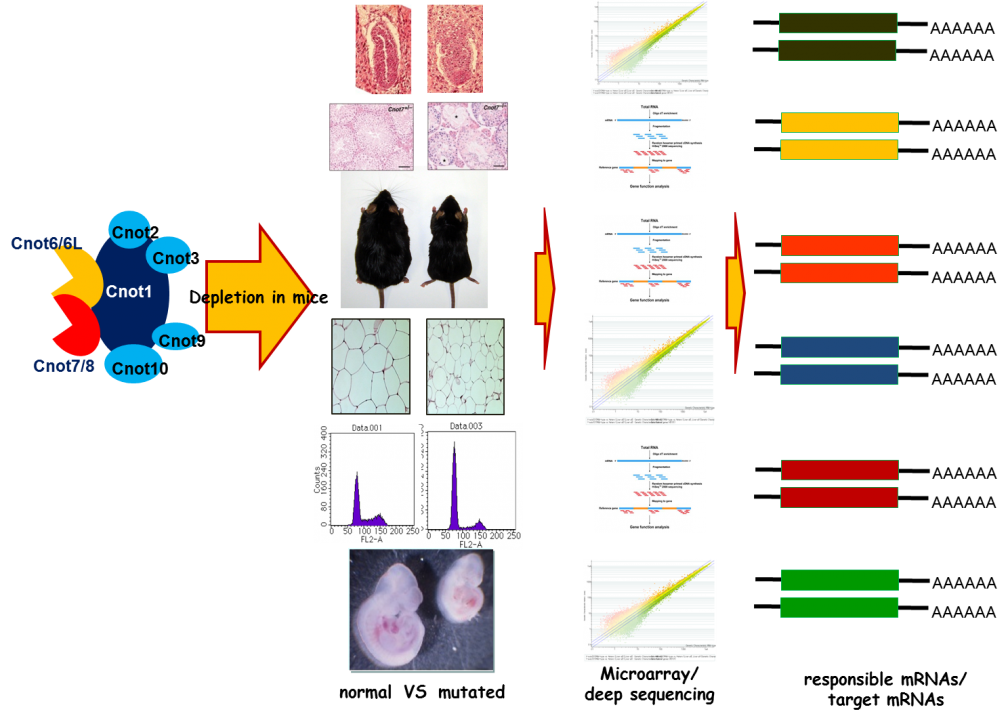
Fig. 4 Strategy for the analysis of CCR4-NOT target mRNAs
2. Roles of protein-kinase signaling and adhesion signaling in the central nervous system.
(i) Fyn tyrosine kinase signaling.
The Src-family protein-tyrosine kinases are implicated in various neural functions such as formation of neural network, myelination, and synaptic plasticity. To analyze the roles of Src and Fyn, we have been focusing on various substrates of these kinases, including N-methyl-D-aspartate (NMDA) type of ionotropic glutamate receptors. Our own studies showed that GluN2A and GluN2B subunits of NMDA receptors, which play important roles in learning, memory formation, and emotional behavior, are the major substrates of Fyn and Src. We identified Tyr-1472 phosphorylation on GluN2B and Tyr-1325 phosphorylation on GluN2A as the major tyrosine phosphorylation site of GluN2B and GluN2A, respectively. Using the knock-in mouse lines expressing mutant GluN2B with a Tyr-1472-Phe (Y1472F) mutation or expressing mutant GluN2A with a Tyr-1325-Phe (Y1325F) mutation, we showed that GluN2B Tyr-1472 phosphorylation is important for fear-related learning in the amygdala and that GluN2A Tyr-1325 regulates depression-related behavior. We further obtained data suggesting that Tyr-1472 phosphorylation of GluN2B is involved in development of postherpetic allodynia due to nerve damage and that the nerve damage at the acute herpetic phase is correlated with the incidence of postherpetic pain.
In parallel of the studies on NMDAR phosphorylation, we have been trying to identify binding partners and substrates of Fyn in the brain using solid-phase phosphorylation screening, yeast two-hybrid screening, and proteomic approaches. As a result, we have identified a number of putative mediators of Fyn-mediated signaling, including NYAP, FAK, p250GAP, TCGAP, Nogo-A, and RhoGEFs. Recently, we provided evidence suggesting that TCGAP (=ARHGAP33) is associated with schizophirenia: #1, a single nucleotide polymorphism of TCGAP is associated with decreased expression in patients with schizophirenia; #2, tcgap-/- mice show reduced expression of surface TrkB in neuron, impaired spine development, and deficit in formation of spatial working memory, a phenotype associated with schizophirenia.
(ii) Lemur tyrosine kinases (LMTKs) form a family of protein kinases that are predominantly expressed in the central nervous system. Members of this kinase family, LMTK1-3, comprise an N-terminal transmembrane region, a Ser/Thr kinase domain, and a long C-terminal tail region. We have reported that LMTK2 (or BREK) interacts with a motor protein, myosin VI, and is involved in the regulation of endosomal membrane trafficking in cultured cell lines. LMTK1 also regulates formation of the endocytic regulatory compartment, suggesting that control of membrane traffic is a common function of this family of kinases. Although we have reported that Lmtk2/Brek-deficient mice are infertile and that LMTK2 is essential for the late stages of spermatogenesis, it is not clear how endocytic events are relevant to spermatogenesis. We do not know either the physiological functions of the LMTK2/BREK kinase in the central nervous system. Another member of the LMTK family, LMTK3, is highly expressed in the cerebral cortex, cerebellum, and hippocampus. To address LMTK3’s biological characteristics and in vivo function, we first generated mouse mutants deficient for the lmtk3 gene. We found that lmtk3-/- mice exhibits prominent behavioral abnormalities, including locomotor hyperactivity, reduced anxiety, and decreased depression-like behavior, providing the first evidence for a physiological role of the LMTK family in the central nervous system. Furthermore, we provided evidence that LMTK3 is involved in regulation of endocytic trafficking of NMDAR.
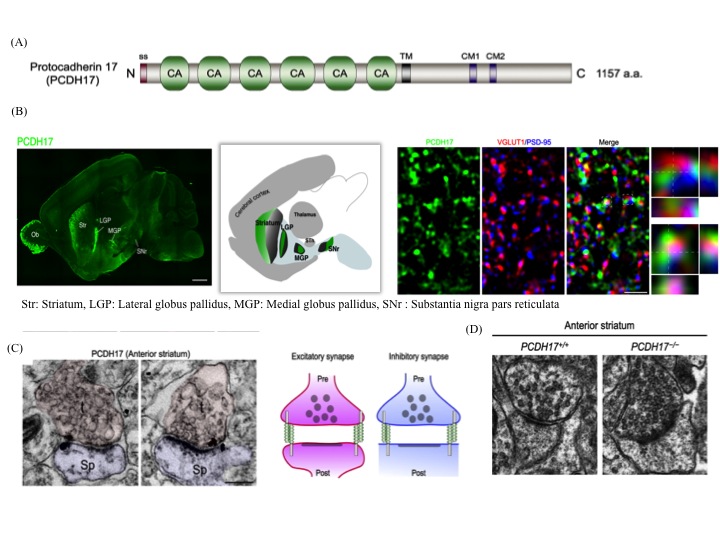
(iii) The anatomical topography of neural circuits generally emphasizes distinct functional units. Functional establishment of this topography requires circuit-specific differentiation and refinement of synapses. Synapse development in individual circuits is organized by various types of adhesion molecules. Proteins of the cadherin superfamily, including the protocadherin family participate in synapse-specific interactions. This family of proteins is expressed in synaptic junctions between different types of neurons in neural circuits. Owing to their highly selective adhesive interactions, cadherin-catenin complexes are required for both pre- and postsynaptic development. Although some cadherin members are expressed in specific zones of the basal ganglia, their roles in circuit-specific synaptic development and their physiological significance remain unclear. We have addressed the biological significance of PCDH17, a non-clustered δ2-protocadehrin family member. Our results indicate that PCDH17 plays a crucial role in the regulation of synaptic vesicle assembly in cortico-basal ganglia circuits. Furthermore, PCDH17 deficiency leads to altered presynaptic function in the cortico-striatal pathway. We also observed antidepressant-like phenotypes in PCDH17−/− mice. These results provide new insights into the mechanisms underlying the synaptic development of specific cortico-basal ganglia circuits and the physiological role of depression-related behaviors.
4. Publications
4.1 Journals
- Delawary, M., Tezuka, T., Kiyama, Y., Yokoyama, K., Wada, E., Wada, K., Manabe, T., Yamamoto, T. & Nakazawa, T. NMDAR2B tyrosine phosphorylation is involved in thermal nociception. Neuroscience letters 516, 270-273, doi:10.1016/j.neulet.2012.04.007 (2012).
- Kohda, K., Kakegawa, W., Matsuda, S., Yamamoto, T., Hirano, H. & Yuzaki, M. The delta2 glutamate receptor gates long-term depression by coordinating interactions between two AMPA receptor phosphorylation sites. Proc Natl Acad Sci U S A 110, E948-957, doi:10.1073/pnas.1218380110 (2013).
- Ohi, K., Hashimoto, R., Nakazawa, T., Okada, T., Yasuda, Y., Yamamori, H., Fukumoto, M., Umeda-Yano, S., Iwase, M., Kazui, H., Yamamoto, T., Kano, M. & Takeda, M. The p250GAP gene is associated with risk for schizophrenia and schizotypal personality traits. PLoS One 7, e35696, doi:10.1371/journal.pone.0035696 (2012).
- Ozaki, Y., Matsui, H., Asou, H., Nagamachi, A., Aki, D., Honda, H., Yasunaga, S., Takihara, Y., Yamamoto, T., Izumi, S., Ohsugi, M. & Inaba, T. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Molecular cell 47, 694-706, doi:10.1016/j.molcel.2012.06.033 (2012).
- Sekino-Suzuki, N., Yuyama, K., Miki, T., Kaneda, M., Suzuki, H., Yamamoto, N., Yamamoto, T., Oneyama, C., Okada, M. & Kasahara, K. Involvement of gangliosides in the process of Cbp/PAG phosphorylation by Lyn in developing cerebellar growth cones. J Neurochem 124, 514-522, doi:10.1111/jnc.12040 (2013).
- Suzuki, T., Tsuzuku, J., Hayashi, A., Shiomi, Y., Iwanari, H., Mochizuki, Y., Hamakubo, T., Kodama, T., Nishitani, H., Masai, H. & Yamamoto, T. Inhibition of DNA damage-induced apoptosis through Cdc7-mediated stabilization of Tob. The Journal of biological chemistry 287, 40256-40265, doi:10.1074/jbc.M112.353805 (2012).
- Takahashi, A., Morita, M., Yokoyama, K., Suzuki, T. & Yamamoto, T. Tob2 inhibits peroxisome proliferator-activated receptor gamma2 expression by sequestering Smads and C/EBPalpha during adipocyte differentiation. Molecular and cellular biology 32, 5067-5077, doi:10.1128/MCB.00610-12 (2012).
- Takeshita, K., Tezuka, T., Isozaki, Y., Yamashita, E., Suzuki, M., Kim, M., Yamanashi, Y., Yamamoto, T. & Nakagawa, A. Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c. Journal of biochemistry 152, 487-495, doi:10.1093/jb/mvs085 (2012).
- Unezaki, S., Sasaki, A., Mabuchi, T., Matsumura, S., Katano, T., Nakazawa, T., Nishio, N., Andoh, T., Yamamoto, T., Nakatsuka, T., Kuraishi, Y. & Ito, S. Involvement of Tyr1472 phosphorylation of NMDA receptor NR2B subunit in postherpetic neuralgia in model mice. Molecular pain 8, 59, doi:10.1186/1744-8069-8-59 (2012).
- Yanagida, S., Taniue, K., Sugimasa, H., Nasu, E., Takeda, Y., Kobayashi, M., Yamamoto, T., Okamoto, A. & Akiyama, T. ASBEL, an ANA/BTG3 antisense transcript required for tumorigenicity of ovarian carcinoma. Scientific reports 3, 1305, doi:10.1038/srep01305 (2013).
- Yoshikawa, S., Kukimoto-Niino, M., Parker, L., Handa, N., Terada, T., Fujimoto, T., Terazawa, Y., Wakiyama, M., Sato, M., Sano, S., Kobayashi, T., Tanaka, T., Chen, L., Liu, Z. J., Wang, B. C., Shirouzu, M., Kawa, S., Semba, K., Yamamoto, T. & Yokoyama, S. Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene 32, 27-38, doi:10.1038/onc.2012.21 (2013).
4.2 Books and other one-time publications
Nothing to report.
4.3 Oral and Poster Presentations
- Inoue, F., Yamada, K., Yamamoto, T., Kishimoto, T. & Ohsugi, M. Male pronuclear formation in a cell-free system from mouse oocytes, in The 35th Annual Meeting of the Molecular Biology Society of Japan, Fukuoka International Congress Center / Marine Messe Fukuoka (2012).
- Morita, M., Siddiqui, N., Yamamoto, T. & Sonenberg, N. A Critical Role for Deadenylase-mediated mRNA Degradation in Metabolism and Obesity, in The 35th Annual Meeting of the Molecular Biology Society of Japan, Fukuoka International Congress Center / Marine Messe Fukuoka (2012).
- Takahashi, A., Suzuki, T. & Yamamoto, T. Involvement of CCR4-NOT deadenylase complex in the regulation of energy metabolism, in The 85th Annual Meeting of the Japanese Biochemical Society, Fukuoka International Congress Center (2012).
- Yamamoto, T. Cell signaling in the braib, in 19th East Asia Joint Symposium on Biomedical Research August 22nd - 25th, 2012"Molecular Understandings for Physiology and Pathology, Mock-am Hall (Bldg.500), Seoul National University, Seoul, Korea (2012).
- Morita, M., Siddiqui, N., Sonenberg, N. & Yamamoto, T. A Critical Role for Deadenylase-mediated mRNA Degradation in Metabolism and Obesity, in The 35th Annual Meeting of the Molecular Biology Society of Japan, Fukuoka International Congress Center / Marine Messe Fukuoka (2012).
- Ohashi, T., Yamamoto, T. & Ohsugi, M. Expression Analysis of γ-tubulin2 in Cancer Cells, in The 35th Annual Meeting of the Molecular Biology Society of Japan, Fukuoka International Congress Center / Marine Messe Fukuoka (2012).
- Soeda, S., Yamada, K., Yamamoto, T. & Ohsugi, M. Regulation of Multifunctional Mitotic Motor Kid/kinesin-10, in The 35th Annual Meeting of the Molecular Biology Society of Japan, Fukuoka International Congress Center / Marine Messe Fukuoka (2012).
- Suzuki, T., Hata, H., Oyama, M. & Yamamoto, T. Effect of Tob deficiency in hepatocellular carcinoma development, in 71st Annual Meeting of the Japanese Cancer Association, Royton Sapporo, Sapporo Geibukan, Sapporo Education and Culture Hall (2012).
- Takahashi, A., Morita, M., Suzuki, T. & Yamamoto, T. The Role of the CCR4-NOT Deadenylase Complex in Obesity, in The 35th Annual Meeting of the Molecular Biology Society of Japan, Fukuoka International Congress Center / Marine Messe Fukuoka (2012).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report.
6. Meetings and Event
6.1 Seminar
- Title: Phosphorylation induced conformation change of Crk and Cbl proteins and its biological implications
- Date: Oct 26, 2012
- Venue: D014
- Speaker: Fuyuhiko Inagaki
- Professor, Hokkaido University Faculty of Advanced Life Science, Frontier Research Center for Post-genome Science and Technology
6.2 Seminar
- Title: My Midkine research ~ Introduction of Midkine as a tumor marker ~
- Date: Oct 31, 2012
- Venue: C015
- Speaker: Shinya Ikematsu
- Professor, Department of Bioresources Engineering, Okinawa National College of Technology
6.3 Seminar
- Title: CCR4-NOT complex is a crucial regulator of heart function and hypertrophic growth
- Date: Dec 10, 2012
- Venue: C615
- Speaker: Keiji Kuba
- Associate Professor, Department of Biological Informatics and Experimental Therapeutics, Akita University Graduate School of Medicine
6.4 Seminar
- Title: Tales of the 3’ poly(A) tail: Mechanisms and regulation of alternative polyadenylation and differential deadenylation
- Date: Dec 13, 2012
- Venue: D014
- Speaker: Ann-Bin Shyu
- Professor and Jesse H. Jones Chair, Department of Biochemistry and Molecular Biology The University of Texas Medical School at Houston
6.5 Seminar
- Title: Structural insights into 20S proteasome activation by human REGg
- Date: March 4, 2013
- Venue: C016
- Speaker: Mark Bartlam
- Professor and Jesse H. Jones Chair, State Key Laboratory of Medicinal Chemical Biology and College of Life Sciences, Nankai University
6.6 Meeting of JST PRESTO(Sakigake) “Design and Control of Cellular Functions”.
- Date: October 2-4, 2012
- Venue: C210
- Organizers: JST PRESTO (Sakigake)
- The Number of Participants: 40
6.7 OIST and IMSUT joint seminar on biomedical sciences for young scientists.
- Date: November 5-7, 2012
- Venue: B250, Auditorium
- Organizers: The Institute of Medical Science, The University of Tokyo
- The Number of Participants: 70
6.8 JST CREST Seminar “Towards Deeper Understanding of Biodynamic Principles”
- Date: December 2-3, 2012
- Venue: C209, B250
- Organizers: JST CREST
- The Number of Participants: 50
6.9 MEXT Innovative Cell Biology by Innovative Technology
- Date: January 12-13, 2013
- Venue: Seaside House, B250
- Organizers: MEXT Innovative Cell Biology by Innovative Technology
- The Number of Participants: 50



