FY2012 Annual Report
Trans-Membrane Trafficking Unit
Associate Professor Fadel Samatey
Abstract
1. Staff
- Dr. Fadel A. Samatey, Associate Professor
- Dr. Clive S. Barker, Researcher
- Dr. Yasuji Kido, Researcher
- Dr. Hideyuki Matsunami, Researcher
- Dr. Vladimir Meshcheryakov, Researcher
- Dr. Young-Ho Yoon, Researcher
- Dr. Paula Bulieris. Technicain
- Irina Meshcheryakova, Technician
- Seiya Kitanobo, Techncian
- Tomoharu Inoue, Technician
- Dr. Yuzuki Manabe, Technician
- Saeko Hedo, Research Administrator
2. Collaborations
- Theme: Expression and purification of the membrane protein complexes
- Type of collaboration: Joint research
- Researchers:
- Professor Keiichi Namba, Graduate School of Frontier Bioscience, Osaka University, Japan
- Professor Katsumi Imada, Department of Macromolecular Science, Graduate School of Science, Osaka University, Japan
- Theme: Molecular dynamic of the bacteria flagellum by neutron scattering
- Type of collaboration: Collaboration
- Researchers:
- Dr. Giuseppe Zaccaï, Institute of Laue-Langevin, Grenoble, France
- Theme: Structural Study of the bacterial Type II secretion system
- Type of collaboration: Joint research
- Researchers:
- Dr. Jean-Rome Voulhoux, CNRS, Institute of Structural Biology and Microbiology, Marseille, France
- Theme: Structural investigation of disordered proteins
- Type of collaboration: Joint research
- Researchers:
- Professor Alla Kostyukova, School of Chemical Engineering and Bioengineering, Washington State University, Pullman, U.S.A.
- Theme: In vitro, in vivo high-yield production of membrane proteins
- Type of collaboration: Collaboration
- Researchers:
- Dr. Bruno Miroux, Institut de Biologie Physico-Chimique, CNRS, Paris, France
- Dr. Jean-Luc Popot, Institut de Biologie Physico-Chimique, CNRS, Paris, France
- Dr. Francesca Zito, Institut de Biologie Physico-Chimique, CNRS, Paris, France
3. Activities and Findings
Cross-Complementation Study of the Flagellar Type III Export Apparatus
The bacterial flagellum is made of about 30 different proteins [1]. It rotates like a propeller to enable the cells to swim. At the base of the flagellum a type III export apparatus secretes the external protein components. Some pathogenic Gram-negative bacteria make type III secretion systems, which use a homologous apparatus. The flagellar export apparatus consists of about six membrane proteins and three soluble proteins. The membrane proteins are highly conserved with a high sequence identity from species to species. For the Salmonella flagellum the membrane proteins are called FlhA, FlhB, FliO, FliP, FliQ and FliR and the soluble proteins are called FliH, FliI, and FliJ. FlhA and FlhB have membrane-spanning N-terminal domains and large C-terminal cytoplasmic domains. Little is known about the function of the membrane domains of these proteins.
We investigated the cross-complementation of FlhB between distantly-related species. We replaced the S. typhimurium flhB gene with the flhB gene from the hyperthermophilic species Aquifex aeolicus or only the trans-membrane encoding region of the flhB gene to create a gene fusion. We sought to understand properties of the cell, which would enable the foreign proteins to function within the Salmonella flagellar system.
Complementation by A. aeolicus FlhB or an A. aeolicus FlhB/S. typhimurium FlhB chimera. The amino acid sequences of A. aeolicus FlhB and S. typhimurium FlhB are 32% identitical. Strains deleted for the flhB gene expressing AquFlhB or AquSalFlhB were inoculated into soft tryptone agar. After 48 hours, a weak zone of swimming was observed for both strains, but suppressor mutant strains with increased motility were produced only for cells synthesizing the AquSalFlhB chimera.
Six suppressor mutant strains were isolated and the suppressor mutations were identified by genome sequence data analysis (Figure 1). Three strains bore missense mutations in the flhA gene that produced substitutions in FlhA at positions A106E, A106V, or L245P. These mutations are all within the N-terminal trans-membrane domain of FlhA. The other three strains bore suppressor mutations in genes encoding enzymes for the biosynthesis of the respiratory chain electron carrier ubiquinone. One mutation was in ubiA, encoding a Q253P substitution in 4-hydroxybenzoate octaprenyltransferase. Another mutation was in ubiE, encoding a G12V substitution in ubiquinone/menaquinone biosynthesis methyltransferase. The final mutation was in ispG, encoding an I207S substitution in 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase.
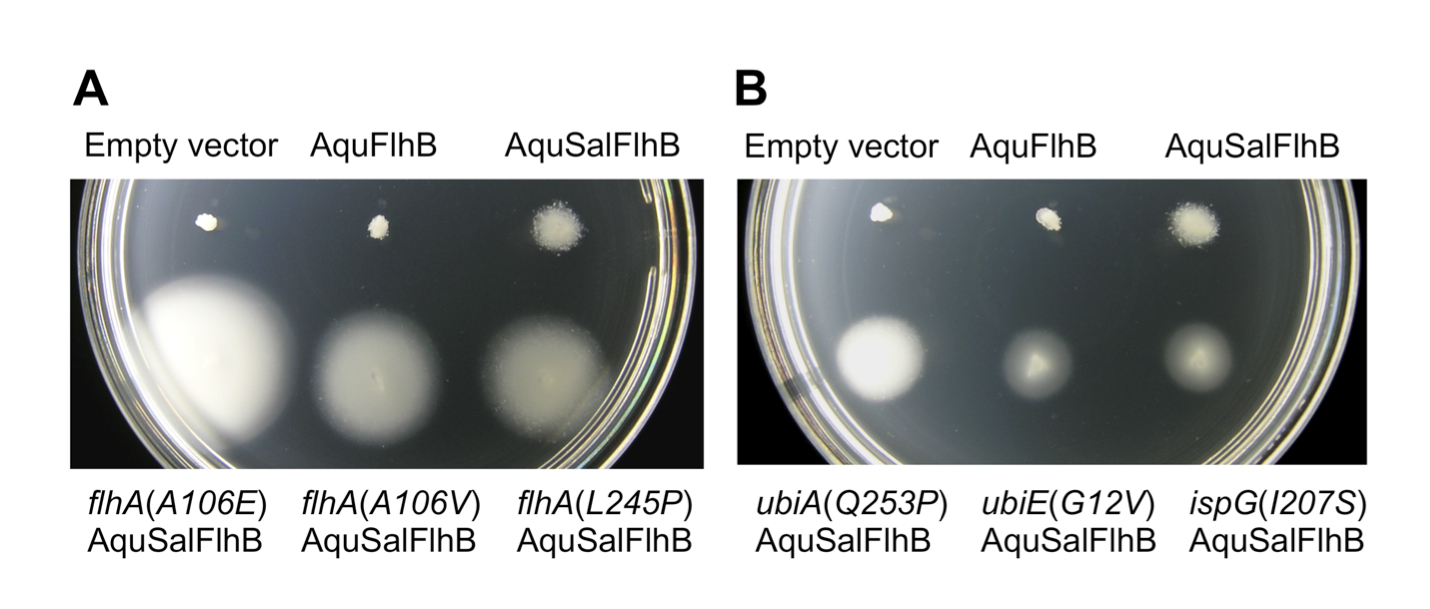 Figure 1. Extragenic suppressor mutations increase motility with an AquSalFlhB chimera. All strains bore an flhB gene deletion. Sal = Salmonella and Aqu = A. aeolicus. (A) Motility phenotype at 24 h of flhB deletion strains harboring either empty plasmid vector, or plasmids synthesizing AquFlhB or an AquSalFlhB chimera. Strains with suppressor mutations in flhA are labeled. The flhB deletion mutant strain complemented with SalFlhB was not included, because it would be overgrown. (B) Motility phenotype at 24 h for flhB deletion mutant strains with suppressor mutations in ubiA, ubiE, or ispG genes and expressing AquSalFlhB. Soft tryptone agar plates were incubated at 30 oC.
Figure 1. Extragenic suppressor mutations increase motility with an AquSalFlhB chimera. All strains bore an flhB gene deletion. Sal = Salmonella and Aqu = A. aeolicus. (A) Motility phenotype at 24 h of flhB deletion strains harboring either empty plasmid vector, or plasmids synthesizing AquFlhB or an AquSalFlhB chimera. Strains with suppressor mutations in flhA are labeled. The flhB deletion mutant strain complemented with SalFlhB was not included, because it would be overgrown. (B) Motility phenotype at 24 h for flhB deletion mutant strains with suppressor mutations in ubiA, ubiE, or ispG genes and expressing AquSalFlhB. Soft tryptone agar plates were incubated at 30 oC.
The ubiA(Q253P), ubiE(G12V) and ispG(I207S) alleles are loss-of-function mutations, which reduced the synthesis of ubiquinone. Reversed-phase high-performance liquid chromatography was used to characterize the quinone pool of the cell membrane of the strains (Table 1). Strains bearing ubiA(Q253P), ubiE(G12V) and ispG(I207S) mutations made reduced ubiquinone. For some ubiA mutant strains, exogenous 4-hydroxybenzoate is known to recover ubiquinone biosynthesis. Accordingly, the strain bearing the ubiA(Q253P) mutation produced 13% of the amount ubiquinone of wild type, but exogenous 4-hydroxybenzoate recovered the amount of ubiquinone to 59%. Conversely, swimming motility of cells bearing the ubiA(Q253P) mutation and synthesizing AquSalFlhB was reduced by adding 4-hydroxybenzoate to the soft tryptone agar (Figure 2).
Table 1. Quantification of the quinone pool of suppressor mutant strains

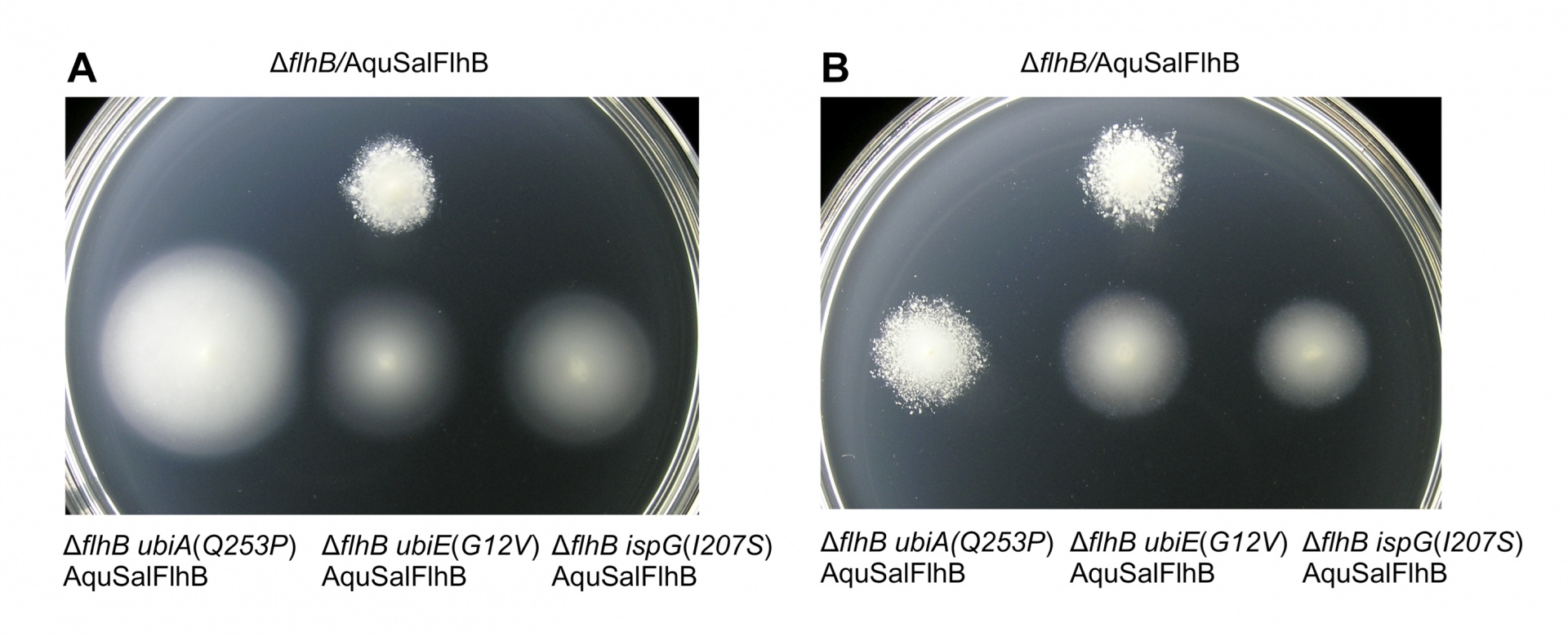 Figure 2. Recovery of ubiquinone biosynthesis for the ubiA(Q253P) mutant reduces complementation of motility by AquSalFlhB. (A) Motility in soft tryptone agar for strains bearing a flhB deletion mutation, or the flhB deletion mutation plus suppressor mutations in the ubiA(Q253P), ubiE(G12V) or ispG(I207S) genes and harboring plasmid expressing the AquSalFlhB chimera. (B) Motility for the same strains as in panel A, inoculated into soft tryptone agar containing 1 mM 4-hydroxybenzoate. The soft tryptone agar plates were incubated for 33 h at 30 oC.
Figure 2. Recovery of ubiquinone biosynthesis for the ubiA(Q253P) mutant reduces complementation of motility by AquSalFlhB. (A) Motility in soft tryptone agar for strains bearing a flhB deletion mutation, or the flhB deletion mutation plus suppressor mutations in the ubiA(Q253P), ubiE(G12V) or ispG(I207S) genes and harboring plasmid expressing the AquSalFlhB chimera. (B) Motility for the same strains as in panel A, inoculated into soft tryptone agar containing 1 mM 4-hydroxybenzoate. The soft tryptone agar plates were incubated for 33 h at 30 oC.
This study shows that despite millions of years of separation between A. aeolicus and S. typhimurium the trans-membrane domains of FlhB have maintained a similar functionality. Suppressor mutations in the flhA gene [flhA(A106E), flhA(A106V) and flhA(L245P)] enabled the Salmonella export apparatus to function better with the AquSalFlhB chimera. The mechanism of these suppressors should rescue how the trans-membrane regions of A. aeolicus FlhB and S. typhimurium FlhB differ most in their interactions with other flagellar components. It is possible that these suppressors could have increased the level of protein-protein interaction between FlhA and FlhB.
The suppressor mutations in the ubiA, ubiE or ispG genes [ubiA(Q253P), ubiE(Q253P), and ispG(I207S)] were surprising; since strains bearing these mutations made reduced levels of ubiquinone it suggests they are loss-of-function mutations. But, despite reduced biosynthesis of ubiquinone these mutations also increased numbers of flagella for cells expressing the AquSalFlhB chimera. However, adding 4-hydroxybenzoate to the growth medium recovered quinone biosynthesis for cells bearing the ubiA(Q253P) mutation, but this also reduced complementation of motility by the AquSalFlhB chimera. This suggests that it was the effect on the quinone pool rather than the mutations in the proteins themselves, which was important in the mechanism of suppression.
Reference
1. Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57: 77-100.
4. Publications
4.1 Journals
- Barker, C. S., Samatey F.A. Cross-complenmentation study of the flagellar type III export apparatus memberane protein FhB. PLoS One Volume 8, e44030.
4.2 Oral and Poster Presentations
- Matsunami, Hideyuki. Molecular basis for the bacterial flagellar P-ring assembly mediated by its periplasmic chaperone, CrSJ Annual Meeting 2012, Miyagi, Japan, Oct 25 -26, 2012
- Meshcheryakov, Vladimir. Crystal structure of the cytoplasmic domain of Salmonella typhimurium FlhB, an essential component of the flagellar secretion system, CrSJ Annual Meeting 2012, Miyagi, Japan, Oct 25 -26, 2012
- Matsunami, Hideyuki. Structural analysis of Salmonella FlgA, an essential chaperone protein for the bacterial flagellar P-ring assembly, AsCA 2012, Adlaide, Australia, Dec 2-5, 2012
- Meshcheryakov, Vladimir. Crystal structure of the cytoplasmic domain of Salmonella typhimurium membrane protein FlhB, an essential component of the flagellar secretion system, AsCA 2012, Adlaide, Australia, Dec 2-5, 2012
- Matsunami, Hideyuki. Conformational Switching of Salmonella FlgA Mediates Chaperone Function for the Flagellar P-ring Assembly, Annual Fragellar Meeting 2012, Gumma, Japan, Mar 3-5, 2013
- Yoon,Young-Ho. X-ray crystallographic structure of the flagellar hook protein from Caulobactor crescentus, Annual Fragellar Meeting 2012, Gumma, Japan, Mar 3-5, 2013
5. Meetings and Events
5.1 Seminar
- Date: November 12, 2012
- Venue: OIST Campus Lab1
- Speaker: Prof. Simon Silver, University of Illinois, Chicago
- Title: "Bacterial Genes (and Enzymes) for all Toxic Metal Resistances"
5.2 Seminar
- Date: November 13, 2012
- Venue: OIST Campus Center Bldg
- Speaker: Prof. Simon Silver, University of Illinois, Chicago
- Title: "Death of the Scientific Journal"
5.3 Lecture
- Date: January 17, 2013
- Venue: OIST Campus Lab1
- Lecturer: Dr. Giuseppe Zaccai, CNRS, Institute de Biology Structurale Institute Laue-Langevin, Grenoble, France
- Title: "Sructural Biology: Small Angles Scattering"
5.4 Lecture
- Date: January 17, 2013
- Venue: OIST Campus Lab1
- Lecturer: Dr. Giuseppe Zaccai, CNRS, Institute de Biology Structurale Institute Laue-Langevin, Grenoble, France
- Title: "Protein Dynamics by inelastic neutorons scattering"
5.5 Seminar
- Date: January 18, 2013
- Venue: OIST Campus Lab1
- Speaker: Dr. Giuseppe Zaccai, CNRS, Institute de Biology Structurale Institute Laue-Langevin, Grenoble, France
- Title: "Life at the extreme edge"
5.6 Seminar
- Date: January 28, 2013 ???
- Venue: Washington State University
- Speaker: Prof. Fadel Samatey, OIST Grauate University
- Title: "???"
5.7 Lecture
- Date: March 14, 2013
- Venue: OIST Campus Lab1
- Speaker: Prof. Alla Kostyukova, Washington State University
- Title: "Protein characterization by CD spectrometer"



