FY2011 Annual Report
Trans-membrane Trafficking Unit
Associate Professor Fadel A. Samatey

Abstract
The flagellum is an organelle that enables Bacteria to swim in their living environment. It is widely spread throughout the bacterial world. Flagella come in different sizes; their length and diameter vary among species. The first observation of a flagellum obtained by electron microscopy in the early 1940’s brought lots of excitement and questions on its structure and how it was being assembled. In bacteria such as Escherichia and Salmonella, the flagellum can be divided in three main parts: the filament, the hook and the basal body. The filament is made by the polymerization of tens of thousands of copies of the flagellin protein (FliC). It is the most visible part of the flagellum. The hook is a short, highly curved segment made by the polymerization of about one hundred copies of the protein, FlgE. The basal body contains different parts of the flagellum: the motor, the switch complex and the rod. The rod connects the hook to the motor. It transmits the torque of the motor directly to the hook. The motor is made of a stator, MotA and MotB, and the rotor, FliG, and the MS ring, FliF. The switch complex, that controls the rotation of the motor clockwise or counter-clockwise, consists of two proteins, FliM and FliN. To complete the description of the bacterial flagellum, some additional parts need to be mentioned. Between the hook and the filament there are two junction proteins named HAP1 (FlgK) and HAP3 (FlgL) (hook-associated proteins 1 and 3) and at the tip of the filament, there is a capping structure made by HAP2 (FliD). The proteins making the rod, the hook, the filament and the junction between the hook and the filament are often referred to as the flagellar axial proteins.
A particular feature in the bacterial flagellum is the existence of a central channel running along the length of the flagellum. Through this channel, the flagellar axial proteins are exported to assemble at the distal end and form the flagellum. The export of these proteins is directed by the flagellar type III export apparatus. In a similar way, pathogenic Gram-negative bacteria also possess an appendage named the Needle Complex (NC) that is used for the secretion of virulence proteins into host cells. This secretion is done by a virulence-associated Type III secretion system (T3SS) that is homologous to export apparatus found in the flagellar system. The T3SS is present in both plant and animal pathogens and is evolutionarily related to the one found in the flagellar system.
In the Trans-membrane Trafficking Unit, we combine microbiology, biochemistry and structural biology to understand 1) the overall structure of the bacterial flagellum and 2) the function of the type III export apparatus.
1. Staff
- Dr. Fadel A. Samatey, Associate professor
- Dr. Clive Barker, Researcher
- Dr. Yasuji Kido, Researcher
- Dr. Hideyuki Matsunami, Researcher
- Dr. Vladimir Meshcheryakov, Researcher
- Dr. Young-Ho Yoon, Researcher
- Dr. Paula Bulieris. Associate Researcher
- Julie Carrere, Technician
- Irina Meshcheryakova, Technician
- Saeko Hedo, Administrative Assistant
2. Collaborations
- Theme: Expression and purification of the membrane protein complexes
- Type of partnership: Joint research
- Researchers
- Professor Keiichi Namba, Graduate School of Frontier Bioscience, Osaka University, Japan
- Professor Katsumi Imada, Department of Macromolecular Science, Graduate School of Science, Osaka University, Japan
- Theme: Molecular dynamic of the bacteria flagellum by neutron scattering
- Type of partnership: Collaboration
- Researcher:
- Dr. Giuseppe Zaccaï, Institute of Laue-Langevin, Grenoble, France
- Theme: Structural Study of the bacterial Type II secretion system
- Type of partnership: Joint research
- Researcher:
- Dr. Jean-Rome Voulhoux, CNRS, Institute of Structural Biology and Microbiology, Marseille, France
- Theme: Structural investigation of disordered proteins
- Type of partenership: Joint research
- Researcher:
- Professor Alla Kostyukova, School of Chemical Engineering and Bioengineering, Washington State University, Pullman, U.S.A.
- Theme: In vitro, in vivo high-yield production of membrane proteins
- Type of partnership: Collaboration
- Researchers:
- Dr. Bruno Miroux, Institut de Biologie Physico-Chimique, CNRS, Paris, France
- Dr. Jean-Luc Popot, Institut de Biologie Physico-Chimique, CNRS, Paris, France
- Dr. Francesca Zito, Institut de Biologie Physico-Chimique, CNRS, Paris, France
3. Activities and Findings
Crystallization of the hook protein, FlgE, from Campylobacter jejuni
a) Introduction
The filament of Salmonella enterica serovar Typhimurium (S. typhimurium) is the one of the most studied and it has been used as a model for the structural and functional studies of bacterial flagella in general. Although the proteins that make all the axial sub-structure of the flagellum come, depending on the bacterium, in a wide range of molecular weights, it has long been accepted that, for all bacteria, both the filament and the hook can be represented as a tubular structure made by the assembly of 11 protofilaments. However, recent study done on the filament of Campylobacter jejuni (C. jejuni) proposed that its filament is made of 7 protofilaments (Galkin et al., 2008). This model would imply that the hook of C. jejuni is either also made of 7 protofilaments because of the way the filament and hook join to make the bacterial flagellum or it has a complete different junction structure compared to S. typhimurium hook-filament junction. To have a better understanding of the flagellum of C. jejuni, and to see the structural and functional difference with the hook of S. typhimurium, we proceeded to crystallize a 79 KDa fragment FlgE2 (91KDa) from C. jejuni strain Cj1729c. This fragment was named FlgE79cj. C. jejuni possesses two hook genes that give two distinct FlgE proteins, FlgE1 (58 KDa) and FlgE2 (92 KDa), that have 27% of sequence identity between them, while having respectively 29% and 33% identity with S. typhimurium FlgE protein. We decided to study FlgE2 instead of FlgE1 because FlgE2, not FlgE1, is known to form the functional hook filament in C. jejuni (Hendrixson & DiRita, 2003). We wanted to see where the extra sequence of FlgE2 would fit when compared to the structure of S. typhimurium FlgE. The 79 KDa fragment of FlgE2-Cj1729c was selected based on its sequence alignment with the 31 KDa fragment of FlgE from S. typhimurium that has been crystallized (Samatey et al., 2004a) and its atomic structure solved by X-ray crystallography (Samatey et al., 2004b) (Fig. 1). We hereby describe the expression, purification and crystallization of native and selenomethionine (SeMet) derivative protein of FlgE79cj and the structure of FlgE79cj, ta major fragment of the hook protein from Campylobacter jejuni.
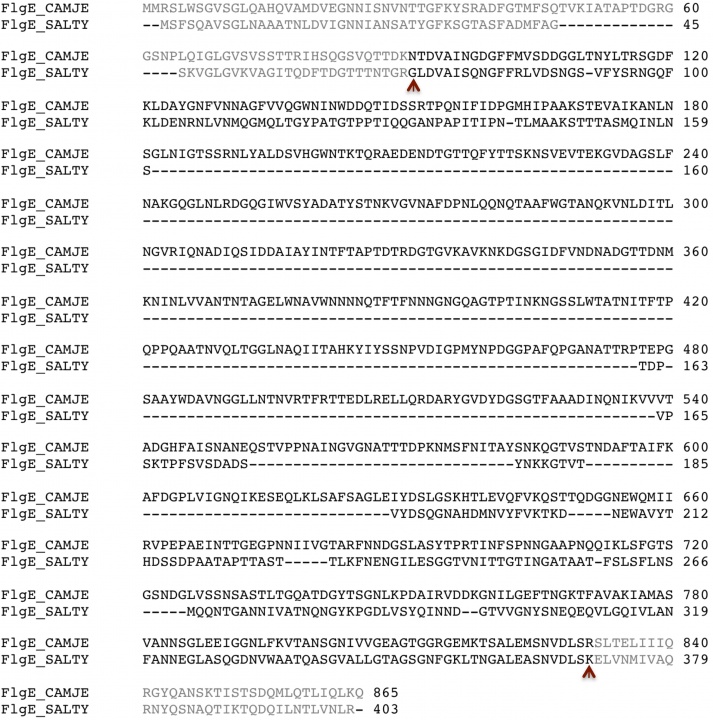
Figure 1. Sequence alignment FlgE from C. jejuni (FlgE_CAMJE) and from S. typhimurium (FlgE_SALTY). The brown arrows mark the beginning and the end of the fragments that were used for crystallization.
b) Cloning expression and purification
The DNA fragment encoding FlgE79cj was amplified by polymerase chain reaction (PCR) from C. jejuni strain NCTC11163. The overexpression construct in pET22b was prepared such that a methionine residue at the N-terminal end and stop codon at the C-terminal end of amino acids 91-831 of the mature polypeptide of Campylobacter jejuni NCTC11166 were added. The forward primer 5'- CAAGCATATGAACACTGACGTTGCTATAAATGGC-3' and the reverse primer 5'-GGTGCTCGAGTTAACGACTTAAATCCACATTTGACATTTC-3' were designed to amplify the gene sequence by the polymerase chain reaction (PCR). The PCR product was ligated into Nde I and Xho I site in pET22b vector. The construct was then transformed into BL21 Rosetta (DE3) (Novagen) and B834 (DE3)/pRARE (Novagen) cells for expression and purification respectively.
A single colony of BL21 Rosetta (DE3) cells harboring Flg79Cj was inoculated in to 50ml of Luria broth medium containing 50 µg ml-1 ampicillin and 34 µg ml-1 chloramphenicol and allowed to grow overnight at 310 K with 275 rpm. The culture was used to inoculate 5L of Luria broth containing antibiotics in a 10L-fermentor. The cells were induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside at the OD600 of 0.5 and the cultivation continued for 3 hours at 310 K. The culture broth was harvested by centrifugation at 8000g for 20min. The cell pellet was suspended in 50 mM NaCl, 20 mM Tris-HCl pH 7.5 and disrupted by sonication. The solution was centrifuged at 10 000g for 15 min to remove the cell debris. The supernatant was centrifuged at 100 000g for 1 hour and the supernatant was loaded onto a Hi-Trap Q-ff column (GE-Healthcare) equilibrated with 20 mM Tris-HCl pH 7.5. Elution was performed with the same buffer solution with a linear gradient of NaCl concentration from 0 to 0.5 M. The main peak fraction was dialyzed against 20 mM Tris-HCl pH 7.5 and loaded onto a Hi-Load Q-Sepharose “High Performance” anion-exchange column (GE-Healthcare) equilibrated with 20 mM Tris-HCl pH 7.5. Elution was performed with a linear gradient of NaCl concentration from 0 to 0.5 M. To enhance the purification, the elution fractions containing the protein were gathered and loaded onto a gel filtration column Superdex-200 (GE-Healthcare) equilibrated with 100 mM NaCl, 10mM Tris-HCl pH 7.5 and eluted with the same solution. The fractions containing the protein were gathered together, dialyzed overnight against 5 mM Tris-HCl pH 7.5. The protein was concentrated to a final concentration of 30 mg ml-1 with a Centriprep centrifugal filter device (Millipore). This protein stock solution was kept at 277 K.
c) Crystallization and data collection
Initial screenings of crystallization conditions were carried out using the sitting-drop vapour-diffusion method. The crystallization drops were initially prepared with an automated nanolitre liquid-handling system (Mosquito, TTP Labtech) in 96-well plates. Three different protein concentrations, 5, 10 and 15 mg ml-1, were used and crystallization plates were prepared by mixing 150 nl protein solution with 150 nl reservoir solution and equilibrated against 120 ml reservoir solution. Crystallization plates were put to equilibrate at 293 K, 288 K and 283 K. For the initial crystallization trials, screening kits from Hampton Research and from Emerald BioSystems were used. Many conditions from the initial screen gave results, mainly at 293 K and 288 K that we thought might be interesting for further crystallization refinements. For most of these conditions, the reservoir solutions contained sulfate salts or polyethylene glycol (PEG) as precipitants agents (Fig. 2a, 2b). Fine-tuning the crystallization of FlgE79cj was done manually using the hanging vapor diffusion method. The PEG precipitant was left out because playing with the PEG molecular weight and concentration did not bring the expected improvements of the crystals shape and size (Fig. 2c, 2d). After extensively screening many sulfate salts as precipitants, we obtained crystals at 288 K in 1.5 M (NH4)2SO4, 10% Isopropanol, 50 mM NaCl, 100 mM CHES, pH 9.5 (Fig. 2e). Unfortunately these crystals dissolved when we tried to pick them up. We decided to leave aside crystallization conditions that used Ammonium Sulfate and to focus on Magnesium Sulfate (MgSO4) and on Dipotassium Phosphate (K2HPO4). Using these two precipitating agents, we obtained crystals after three weeks, in a wide range of buffers with pH varying from 4.5 to 9.5 and with a protein concentration of 15 mg ml-1. The best crystals we obtained at 293 K, in 0.8 M K2HPO4, 0.1 M Acetate pH 4.5, belong to the space group I4122 with the cell parameters a= b= 186.2 Å, c= 386.6 Å, a= b= g= 90°. Unfortunately these crystals did not diffract beyond 5.0 Å at beam line BL44XU of SPring-8 synchrotron. The addition of NaCl and the change of buffer did not bring any positive change. To improve the diffraction limit of the crystals, we decided to combine some additives to the initial crystallization conditions. Dipotassium phosphate combined with 5% isopropanol gave better-looking crystals (Fig. 2f) but there was no improvement in the diffraction limit. Increasing the concentration of isopropanol did not bring better improvements. Isopropanol combined to Magnesium Sulfate did not give better results. However, we found that by adding Cadmium Sulfate to Magnesium Sulfate we obtained crystals that seemed to have different shape (Fig. 2g). We switched to Dimethyl sulfoxide (DMSO) as additive. The addition of less than 10% of DMSO to K2HPO4 left crystals size, shape and diffraction limit unchanged. When the concentration of DMSO in the reservoir was between 10% and 14% (5% and 7% in the drop) relatively thin, long stack crystals were obtained (Fig. 2h). These crystals diffract to about 2.1 Å and belong to space group of P21 and the cell parameters of a= 75.7 Å, b= 173.8 Å, c= 150.8 Å, a= g= 90°, b= 106.5°. SeMet containing crystals of FlgE79cj were obtained in similar conditions. The data collection statistics are summarized in Table 1.
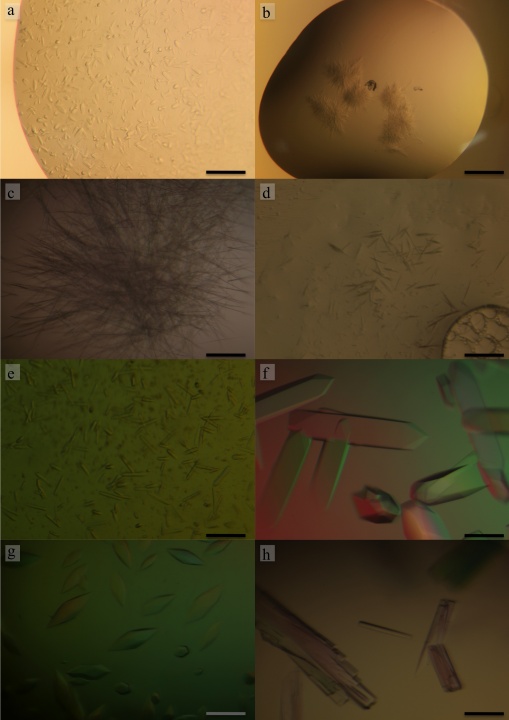
Figure 2. Crystals of FlgE79cj from C. jejuni obtained in: a) in 1.5 M (NH4)2SO4, 0.1 M Tris-HCl pH 8.5, b) 30% PEG 3k, 0.1 M CHES pH 9.5, c) 10% PEG-10K, 4% Dioxane, 0.1 M Citrate pH 5.5, d) 30% PEG-400, 0.1 M CHES pH 9.5, e) 1.5 M (NH4)2SO4, 50 mM NaCl, 10% Isopropanol, 0.1 M CHES pH 9.5, f) 0.8 M K2HPO4, 5% Isopropanol, Acetate 0.1 M pH 4.5 g) 1.4 M MgSO4, 50 mM NaCl, 6 mM CdSO4, Tris-HCl pH 8.5, h) 1.2 M K2HPO4, 50 mM NaCl, 5 mM Tl2SO4, 12% DMSO, 50 mM MES, pH 6.0. The scale bar on the bottom right represents 0.1 mm.
d) Results and discussion
Diffraction data sets were collected at SPring-8, on Beamline BL44xu for the SeMet crystal and on beamline BL41Xu for the native data (fig. 3). SeMet data was taken at SPring-8 on beamline BL44xu. Data sets were processed with MOSFLM (Leslie, 1992) and scaled with SCALA (Evans, 2006) from CCP4 software suite (Winn et al., 2011). Data reduction and systematic absences of reflections indicated that FlgE79cj crystals obtained in DMSO belong to space group P21. The Matthews coefficient (Matthews, 1968) VM was calculated to be 3.0 Å3Da-1 for 4 molecules per asymmetric unit and 2.4 Å3Da-1 for 5 molecules per asymmetric unit. These values correspond to a solvent content of 58% and 48%, respectively. Using the Phenix software package (Adams et al., 2010), the program Phenix-AutoSol found 42 SeMet sites. Few of the SeMet found had a very low occupancy. FlgE79cj contains 10 Met (including the N-terminal methionine). The initial model building calculations, on the basis of 4 and 5 molecules per asymmetric unit, were done using the program Phenix.AutoBuild. These preliminary calculations showed that the asymmetric unit contains 4 molecules. The structure of FlgE79cj is now in the last stage of refinement.
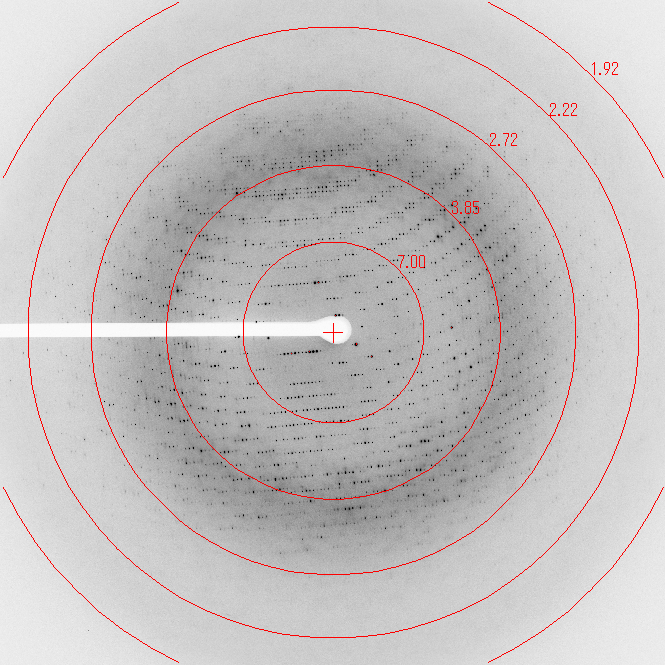
Figure 3. Diffraction pattern from the native crystal of FlgE79cj
The difficulties in the crystallization of FlgE79cj were due to the fact that early crystallization trials gave us many hits. It was difficult to refine all these initially interesting conditions. The crystal used to collect the native data were obtained from a solution initially containing 1.2 M K2HPO4, 50 mM NaCl, 5 mM Tl2SO4, 12% DMSO, 50 mM MES, pH 6.0. Although XAF measurements did not confirm the presence of thallium in the crystal, crystals grown with thallium tended to diffract better than those without. The SeMet crystal used to collect the MAD data grew in a solution of K2HPO4 1.4M, NaCl 50 mM, DMSO 14%, Citrate 0.1 M pH 5.5 with a protein concentration of 15 mg ml-1.
Many of the early crystals, which belong to the space group I4122, were big enough to be checked, at 100 K, on our laboratory X-ray source, a Rigaku FR-E+, these crystals diffracted poorly in spite of a long exposure time. The need to check the crystal on a synchrotron beamline slowed down the project and prevented an extensive screen of both pH and salt additives when using DMSO. As it often happens in protein crystallization, the nice looking crystals are not always the best diffracting.
e) References
Adams, P. D. et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst.D. 66, 213-221.
Berg, H. C. & Anderson, R. A. (1973). Bacteria swim by rotating their flagellar filaments. Nature. 245, 380-382.
Berg, H. C. (2003). The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54.
DePamphilis, M. L. & Adler, J. (1971). Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J. Bacteriol. 105, 384–395.
Evans, P. (2006). Scaling and assessment of data quality. Acta Cryst.D. 62, 72-82.
Galkin, V. E., Yu, X., Bielnicki, J., Heuser, J., Ewing, C. P., Guerry, P. & Egelman, E., H. (2008). Divergence of quaternary structures among bacterial flagellar filaments. Science. 320, 382-385.
Hendrixson, D. R. & DiRita, V. J. (2003). Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50, 687–702.
Homma, M., DeRosier, D. J. & Macnab, R. M. (1990). Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J. Mol. Biol. 213, 819-832
Leslie, A.G. (2006). The integration of macromolecular diffraction data. Acta Cryst. D. 62, 48-57.
Macnab, R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100.
Matthews, B. W. (1968). Solvent content of protein crystals. J. Mol. Biol. 33, 491–497.
Namba, K. & Vonderviszt, F. (1997). Molecular architecture of bacterial flagellum. Q. Rev. Biophys. 30, 1-65.
Samatey, F. A., Matsunami, H., Imada, K., Nagashima, S. & Namba, K. (2004a). Crystallization of a core fragment of the flagellar hook protein FlgE Acta Cryst. D. 60, 2078-2080
Samatey, F. A., Matsunami, H., Imada K, Nagashima, S., Shaikh, T. R., Thomas, D. R., Chen, J. Z., DeRosier D. J., Kitao, A. & Namba, K. (2004b). Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature. 431, 1062-1068.
Winn, M. D. et al. (2011). Overview of the CCP4 suite and current developments. Acta Cryst. D67, 235–242.
4. Publications
4.1 Journals
- Matsunami H., Samatey F.A., Nagashima S., Imada K. and Namba K. (2012). Crystallization and preliminary X-ray analysis of FlgA, a periplasmic protein essential for flagellar P-ring assembly. Acta Crystallogr. Section F 68,310-3.
- Kido Y., Yoon Y.H. and Samatey F.A. (2011). Crystallization of a 79 kDa fragment of the hook protein FlgE from Campylobacter jejuni. Acta Crystallogr. Section F 67,1653-7.
- Meshcheryakov V.A., Krieger I., Kostyukova A.S. and Samatey F.A. (2011). Structure of a tropomyosin N-terminal fragment at 0.98 Å resolution. Acta Crystallogr. Section D 67, 822-5.
- Meshcheryakov V.A. and Samatey F.A. (2011). Purification, crystallization and preliminary X-ray crystallographic analysis of the C-terminal cytoplasmic domain of FlhB from Salmonella typhimurium. Acta Crystallogr. Section F 67, 808-11.
4.2 Oral and Poster Presentations
- Clive Barker, Coupling of Type III secretion to the membrane potential, Biophysical Society 56th Annual Meeting, in San Diego, CA, Feb 25-29, 2012
- Vladimir Mshcheryakov, Structural flexibility of the cytoplasmic domain of flagellar type III secretion protein FlhB is important for the function of the protein, Biophysical Society 56th Annual Meeting, in San Diego, CA, Feb 25-29, 2012
5. Meetings and Events
5.1 Workshop: Practical Protein 3D Structure Visualization and Structural Bioinformatics
- Date: May 22 - 23, 2011
- Venue: OIST Campus
- Speaker: Prof. Eric Martz, University of Massachusetts, Amherst (U.S.A.)
5.2 Seminar: Out of Africa: DNA genomic data showing the origin of humankind and subsequent global migration
- Date: November 2, 2011
- Venue: OIST Campus
- Speakers: Prof. Simon Silver, University of Illinois at Chicago (U.S.A.)
5.3 Seminar: Formation of 2NaBH4/MgH2 system from 2NaH/MgB2 by hydrogenation
- Date: December 12, 2011
- Venue: OIST Campus
- Speakers: Dr. Maria Orlova, European Synchrotron Radiation Facility, Grenoble (France)
5.4 Workshop: OIST/CCP4 School 2011
- Date: December 5-9, 2011
- Venue: OIST Seaside House
- Co-organizers:
- Dr. Charles Ballard, STFC Rutherford Appleton Laboratory (UK)
- Dr. Ulf Skoglund, Okinawa Institute of Science and Technology (Japan)
- Speakers:
- Dr. Charles Ballard (CCP4/STFC, Oxford, UK)
- Dr. Ciaran Carolan (EMBL, Hamburg, Germany)
- Dr. Phil Evans (MRC-LMB, Cambridge, UK)
- Dr. Kazuya Hasegawa (Spring-8, Japan)
- Dr. Ronan Keegan (CCP4/STFC, Oxford, UK )
- Dr. Eugene Krissinel (CCP4/STFC, Oxford, UK)
- Dr. Victor Lamzin (EMBL, Hamburg, Germany)
- Dr. Bernhard Lohkamp (Karolinska Inst., Stockholm, Sweden)
- Dr. Garib Murshudov (MRC-LMB, Cambridge, UK)
- Dr. Nobutoshi Ito (Tokyo Medical and Dental Univ., Japan)
- Dr. Andrea Thorn (Goettingen Univ., Germany)
- Dr. Min Yao (Hokkaido Univ., Sapporo, Japan
- Dr. Xia-Dong Su (Peking University, China )
6. Other
During the fiscal year 2011, the coordinates of the following three-dimension protein structure has been deposited into the Protein Data Bank (PDB):
PDB-id: 3AZD “Crystal structure of tropomyosin N-terminal fragment at 0.98Å resolution”



