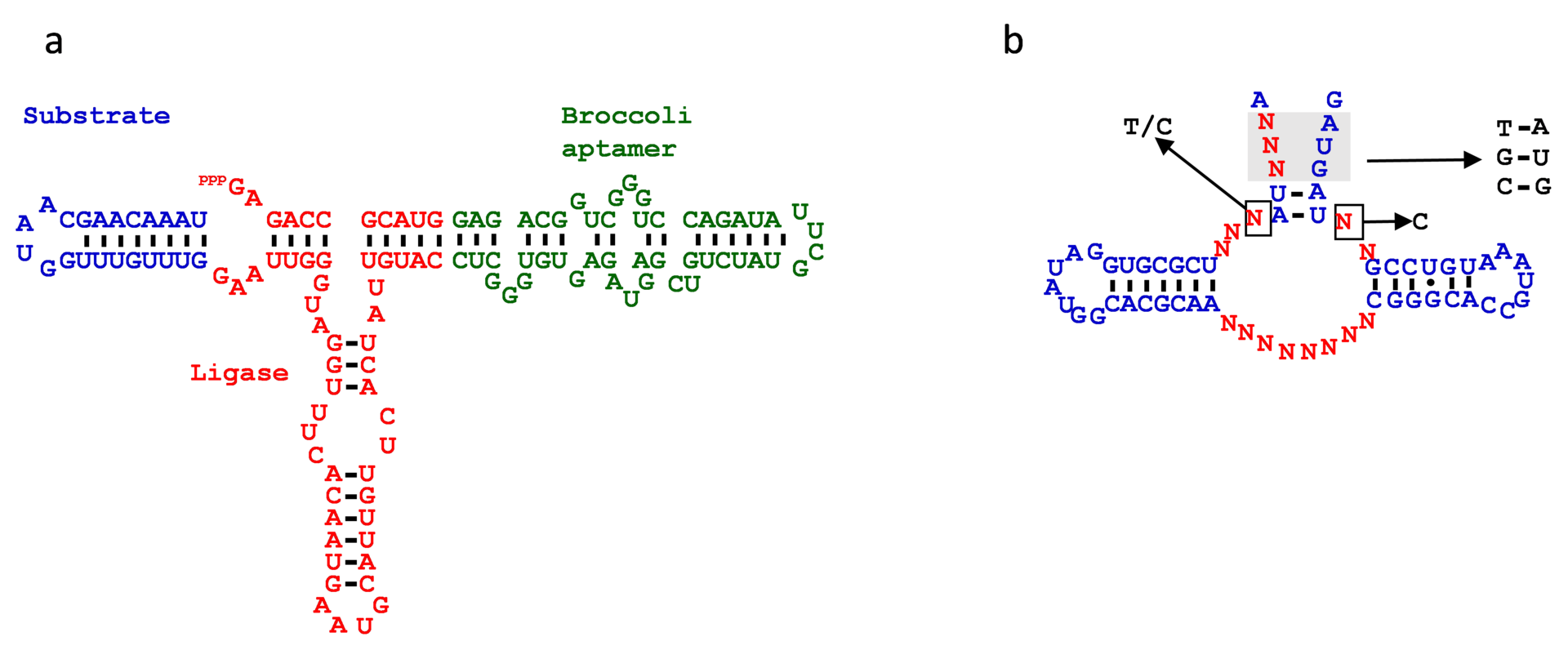Annual Report FY2015: Yoko Nomura
Yoko Nomura
Science and Technology Associate
1. Introduction
I started working at OIST since July 2015. My initial task was to set up my working/research environment within the Nucleic Acid Chemistry and Engineering (Yokobayashi) Unit which was also new to OIST. Subsequently, I started working on several projects that involve engineering of catalytic RNAs in collaboration the Yokobayashi Unit. In addition, I have been developing an independent project on functionalization of silk fiber materials. All of my projects are based on my strong background in biotechnology.
2. Activities and Findings
Engineering of catalytic RNAs (ribozymes)
Some noncoding RNAs have catalytic activities and are involved in regulating gene expression in cells. Researchers have also discovered artificial RNAs with catalytic activities by in vitro selection. These catalytic RNAs, or ribozymes, can be and have been engineered for various potential applications in biotechnology and medicine.
1) Self-circularizing RNA ligase
Biologists have recently discovered a number of circular RNAs in diverse organisms. While their biological functions remain largely unknown, circular RNAs are expected to have unique properties compared the more common linear RNAs, for example, increased stability inside the cell. We developed a fully artificial mechanism to generate circular RNAs in vitro by modifying a compact RNA ligase ribozyme discovered by the Joyce group [1] (Figure 1a). To demonstrate that the circular RNA can be equipped with another functional RNA module, we inserted the Broccoli aptamer that can bind to a nonfluorescent small molecule to emit green fluorescence [2]. We confirmed that the RNA circularized by gel electrophoresis and by selective digestion of linear RNA by RNase R. However initial results indicate that the circularized RNA is unstable under the reaction conditions, and is slowly and irreversibly linearized. Additional modification of the ribozyme sequence and/or reaction conditions are being considered for further investigations.
2) Scaffold-based aptamer selection and screening
RNA aptamers are RNA sequences that can specifically bind targeted molecules. Aptamers are typically obtained by in vitro selection (SELEX) from >1013 random sequences. However, the conventional aptamer selection process is labor intensive and time consuming. Another challenge when isolating RNA aptamers that bind to small molecules is the need to immobilize the target molecule. We are trying to address these challenges by exploiting the next-generation sequencing (NGS) technology available at OIST. Our strategy is to exploit a natural RNA aptamer that binds guanine as a scaffold, and isolate aptamers capable of binding different molecules by varying specific nucleotides that are more likely to directly contact the targeted molecule.
Towards this goal, we designed and constructed a large RNA library based on the guanine aptamer scaffold with 16 randomized nucleotides (4.3 X 109 mutants) (Figure 1b) fused to an HDV ribozyme. We performed 5 rounds of selection for binding affinity to guanine and analyzed the resulting RNA population by NGS. We confirmed that some of the randomized bases converged to specific nucleotides, indicating that the aptamers have evolved to bind the targeted molecule.
|
Figure 1. Engineered RNA sequences. a) Self-circularizing ribozyme equipped with a Broccoli aptamer (green). The red bases comprise the ligase ribozyme developed by Joyce et al. The blue sequence represents the ligase substrate which was covalently linked to the ligase in this construct. b) Guanine aptamer-based library used in this study. The red “N” nucleotides indicate the randomized positions within the natural guanine aptamer scaffold. The bases show in black are the bases that were enriched after in vitro selection. |
Functionalization of silk fiber
As an independent project, I started to work on functionalization of silk fiber.
Silk fiber is an attractive material for biomedical applications as well as for other general applications because of its superior biocompatibility and biodegradability. To make silk fiber materials safer and more valuable, functionalization of silk, such as addition of antibacterial property would be desirable. Chemical and genetic modifications of silk have been reported but these processes are complex and time consuming.
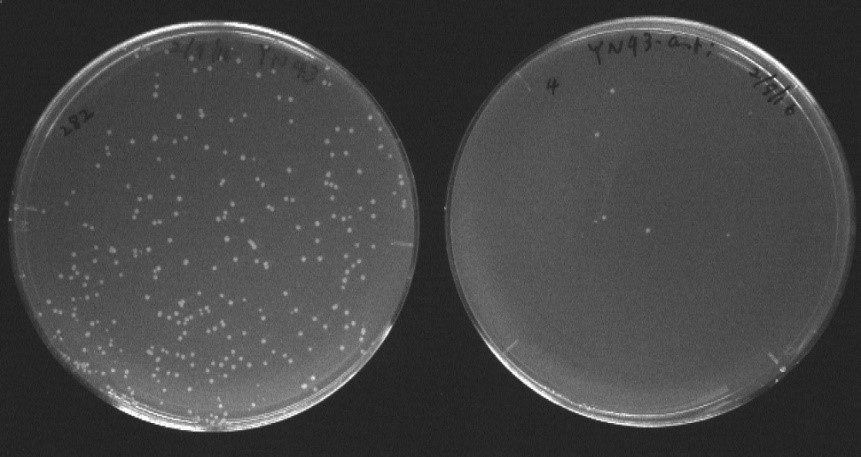
We previously discovered a peptide motif (QSWS) that binds to silk fibroin fiber (Y. Nomura et al, Biotechnol Lett, 33, 1069-73, (2011)) which can be used to impart new functions to silk fiber. I started to develop antibacterial silk fiber using this silk-binding motif fused to antimicrobial peptides (AMP) which are a new class of antibacterial reagents with low occurrences of bacterial resistance. This year, I also submitted a research proposal on this topic for the Kakenhi grant. Result of the feasibility study is shown in Figure 2. A novel fusion peptide (fibroin binding peptide-spacer-AMP) was shown to function as an antibacterial peptide.
|
Figure 2. Antibacterial effect of the novel silk-binding and antibacterial peptide fusion. LB plates inoculated with E. coli on untreated (left, 282 cfu) and peptide-treated (right, 4 cfu) media. |
3. Collaborations
Yokobayashi Unit
4. Publications and other output
S. Kobori, Y. Nomura, A. Miu, Y. Yokobayashi, High-throughput assay and engineering of self-cleaving ribozymes by sequencing, Nucleic Acids Research,43, e85, (2015).
Y. Nomura and Y. Yokobayashi, Aptazyme-based riboswitches and logic gates in mammalian Cells, Methods in Molecular Biology, 1316, 141-148, (2015). (book chapter)
References
1. M. P. Robertson, G. F. Joyce, Chem Biol, 21, 238-245, (2014).
2. G. S. Filonov, J. D. Moon, N. Svensen, S. R. Jaffrey, J Am Chem Soc, 136, 16299-16308, (2014).