Annual Report FY2015: Garth Ilsley
Garth Ilsley
Science and Technology Associate
1. Introduction
The past year has led to the completion and publication of modelling work in Drosophila, as well as the completion of single-cell RNA-seq analysis in Ciona. In addition, with the Luscombe Unit, we have established a stable culture of Oikopleura dioca at OIST.
2. Activities and Findings
1. Modelling transcriptional enhancer function in Drosophila
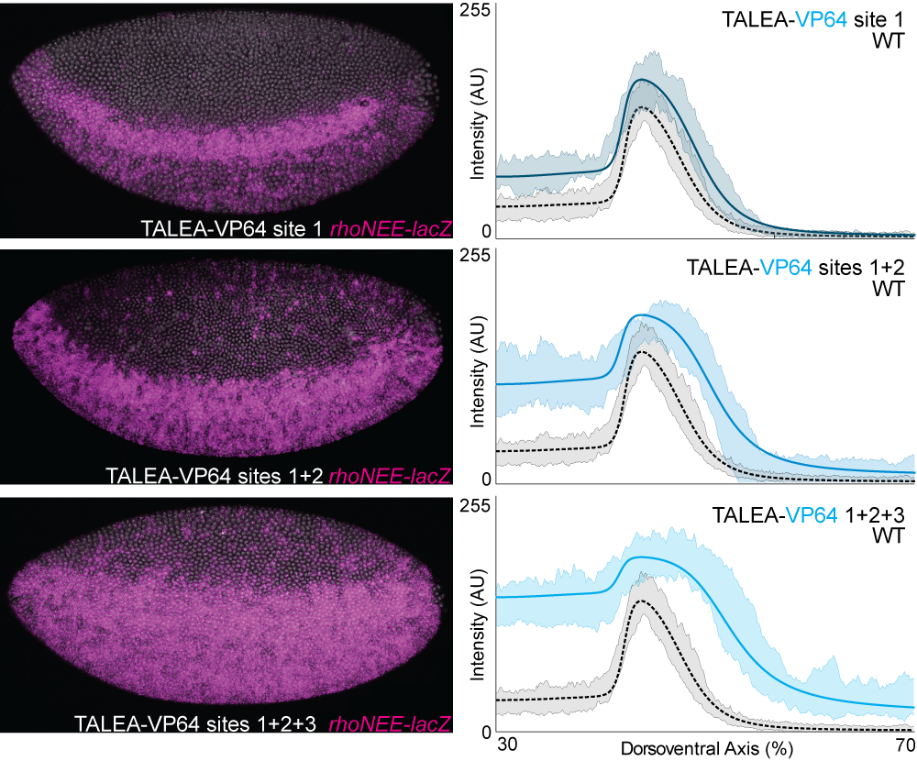
In collaboration with Justin Crocker and David Stern, we have completed and published our analysis of the regulatory function of even-skipped and rhomboid enhancers in Drosophila. We validated the results of model predictions of enhancer function with experimental perturbations, particularly those using transcription activator–like effectors fused to activators or repressors (TALEAs and TALERs).
2. Single-cell measurements in Ciona intestinalis
It is important to have measurements of transcription factor levels and gene outputs at single-cell resolution. Spatially averaging transcription factor measurements obscures cellular state and hence limits the effectiveness of statistical studies of the regulatory inputs and outputs in the cell. Therefore, to understand gene regulation it is crucial to develop approaches for measuring gene expression at the single cell level.
I have finished RNA-seq analysis and generated a complete dataset of single-cell expression data at a specific stage of development in Ciona, a marine chordate. Experimental validation work by my collaborators in the Luscombe Lab is continuing.
3. Establishment of Oikopleura dioica culture
Over the past few years, the Luscombe Unit has developed expertise and knowledge working with Ciona, an invertebrate marine chordate. Future experimental work will benefit from having an organism similar to Ciona, but with a shorter lifecycle. Oikopleura dioica is such an animal. With the help of the lab of Hiroki Nishida at Osaka University we have established a reliable culture of Oikopleura dioca at OIST.
3. Collaborations
Theme: Measuring gene expression with single-cell RNA-seq
Type of collaboration: Joint Research
Researchers: Luscombe Unit (OIST)
Theme: Establishing Oikopleura dioica culture at OIST in the Luscombe Lab
Type of collaboration: Assistance and guidance from experts in the field
Researchers: Takeshi Onuma, Tatsuya Omotezako and Hiroki Nishida (Osaka University)
Theme: Modelling transcriptional enhancer function in Drosophila
Type of collaboration: Joint Research
Researchers: Justin Crocker and David Stern (Janelia Research Campus, USA)
4. Publications and other output
Crocker, J., Ilsley, G. R. & Stern, D. L. Quantitatively predictable control of Drosophila transcriptional enhancers in vivo with engineered transcription factors. Nat Genet 48, 292–298 (2016).



