Annual Report FY2015: Cecilia S. Lu
Cecilia S. Lu
Science and Technology Associate
1. Introduction
Synaptic homeostasis is the hallmark of a healthy nervous system which is capable of constraining hyper-excited neural circuit while boosting underdeveloped neural connections to produce consistent behaviors. Previously, I discovered a neural activity-inducible and conserved microRNA-8 (miR-8) that plays functional roles during neuromuscular synapse development in Drosophila and identified a subset of synaptic cell adhesions molecules (CAMs) mediating downstream effects of miR-8 on both sides of the neuromuscular junctions (NMJ) (Lu et al., 2014, Philos Trans R Soc Lond B Biol Sci. 369(1652). pii: 201305172014). In the past year, I surveyed the protein-protein interaction (PPI) network of one of these synapse-organizing CAMs to identify and investigate molecules recruited for structural homeostasis at the NMJ. In addition, I initiated and established the most technically challenging and necessary parts of experimental protocols towards isolating specific sub-types of neurons for microRNA expression profiling for a new Van Vactor Unit project as Group Leader from 2016.
2. Activities and Findings
2.1 PPI Network of Synaptic CAM Fasciclin 3 (Fas3)
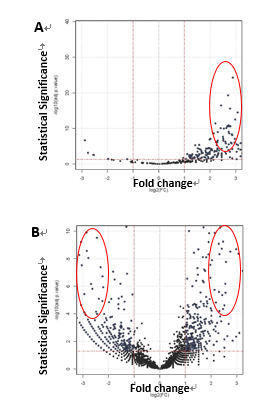
To survey the protein interactome of Fas3 which acts downstream of miR-8 and is known for its role in motor axon targeting and synapse formation, I immunoprecipitated a transgenic epitope-tagged Fas3 expressing selectively in embryonic brain or muscle, followed by tandem LC-Mass Spectrometry. To enrich the recovery of protein complexes from plasma membrane fractions, such as cell surface receptors or ligands, I used a reversible chemical cross-linker. In parallel, I performed antibody pull-down of cytoplasmic protein complexes with high-affinity epitope binding magnetic beads as input for mass spectrometry. In summary, ~2300 proteins were positively detected with confidence in the Drosophila embryonic lysate by mass spectrometry. 28 out of 123 proteins isolated from neuronal Fas3 complexes (Fig. 1A) and 36 out of 185 proteins were short-listed as candidates (Fig. 1B) based on fold-change, statistical and functional significance. A screening panel of 15 available antibodies for immunoprecipitation and Western blotting were used to validate the interaction between Fas3 and candidate independently from the result from mass spectrometry. 6 final candidates (2 CAMs, 1 Toll receptor, 2 Golgi/ER/endosomal transport, 1 Wnt signaling) were confirmed and genetic epistasis experiments to characterize in vivo functions at NMJs were underway.
|
Figure 1. Fas3-interacting proteins identified by affinity purification and tandem liquid chromatography mass spectrometry. A) from epitope-tagged neuronal Fas3 protein complexes. B) from epitope-tagged muscle Fas3 protein complexes. Candidates subjected to secondary screens are encircled. |
2.2 Interaction between Synaptic CAMs Fasciclin 3 (Fas3) and Neuroglian (Nrg) Promotes Structural Homeostasis of Neuromuscular Synapse
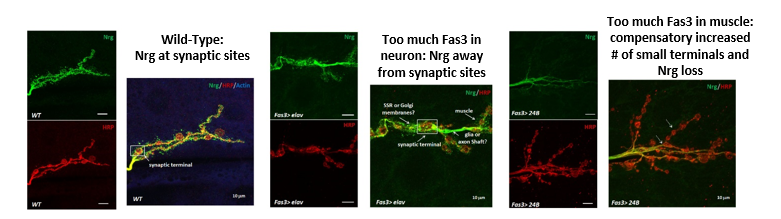
Genetic interaction between Fas3 and Nrg downstream of miR-8 was previously demonstrated in embryonic NMJs (Lu et al., 2014). However, it was not known whether these two synaptic CAMs physically interact with each other and whether the structural homeostasis of synapse is affected by this interaction. Through PPI network analysis and candidate-specific secondary screening (see 2.1), I confirmed that Fas3 and Nrg biochemically interact. Furthermore, I found that increasing Fas3 expression and saturating interactions with Nrg appeared to cause structural changes and mis-localization of neuronal Nrg, moving it away from synaptic terminals into extra-synaptic regions and even into subsynaptic reticulum (SSR) in the muscle or degraded (Fig. 2).
|
Figure 2. Synaptic structure and localization of Neuroglian at neuromuscular synaptic terminals is severely disrupted by mis-expression of Fasciclin 3. HRP is an axonal membrane marker (red), Nrg is probed with neuronal specific isoform of anti-Nrg (green), and Actin is muscle marker (blue). Color-merged images are derived from corresponding single channel images shown to their left. |
2.3 Establishment of Primary Neuronal Culture from Drosophila Embryos
I established and optimized an experimental protocol to culture neurons from dissociated Drosophila embryos to single cells in resuspension and imaged nascent axonal process and synaptic proteins for up to 7 days in culture.
2.4 MicroRNA Expression Profiling of Drosophila Embryonic Motor Neurons
I established and optimized experimental protocols to isolate motor neurons (4% of total dissociated cells) and interneurons (0.1-0.2% of total dissociated cells) by using fluorescence activated cell sorting (FACS). I successfully extracted and enriched small RNAs from low input (100 ng, 10,000 cells) and acquired miRNA expression profile using PCR-free digital barcoding.
3. Collaborations
3.1 Dr. David Van Vactor, Visiting Professor, Formation and Regulation of Neuronal Connectivity Unit
3.2 Dr. Hiroshi Kohsaka, Assistant Professor, University of Tokyo supplied transgenic fly stocks expressing GFP-positive interneurons
4. Publications and other output
4.1 Peer-reviewed Journals - None to report
4.2 Oral and Poster Presentations
OIST STG Forum - Title: Understanding microRNA codes in the development of neural connectivity (November, 2015)
OIST Internal Seminar - Title: Understanding microRNA codes in the development of neural connectivity (March, 2016)
4.3 Teaching Activities
Tutor for OIST Developmental Neurobiology Course 2015