FY2012 Annual Report
Biological Systems Unit
Adjunct Professor Igor Goryanin

Abstract
The main focus of our research is developing the microbial fuel cells. Starting with the lab-scale systems we moved toward the practical application by scaling-up the reactors and developing systems integration.
1. Staff
- Dr. Igor Goryanin, Professor (adjunct)
- Dr. Larisa Kiseleva, Researcher
- Dr. Alexander Mazein, Researcher
- David Simpson, Technical staff
- Midori Tanahara, Research Administrator
2. Collaborations
- Theme: Biological network analysis
- Type of collaboration: Research Collaboration
- Researchers:
- Dr. Tong Hao, Tianjin University
3. Activities and Findings
3.1 Investigation of scaled-up microbial fuel cells in collaboration with awamori distillery for treatment of waste water
Two pilot scale microbial fuel cell systems (MFC, 60L internal volume each) were constructed and tested at OIST for current generation and chemical oxigen demand (COD) removal with awamori distillery waste water. The design of the MFC used continuous flow, and comprised of 8 electrode pairs separated by plastic flow guides. The cathode uses a semi-wet, air circulating design. The pilot began in 6th February and concluded March 31st, a total of 53 days.
The inoculation material consisted of local waste water treatment sludge, which was enriched using locally sourced wild type microorganisms. The enhancement of the internal biofilm to encourage exoelectrogenic activity and COD removal was carried out over several weeks. An average SCOD removal rate of 60% was achieved at organic loading rate (OLR) of 13,000mg COD and a four day hydraulic retention time. Periodically, biological samples were taken from biofilm formations and within the channels of the MFC. These will be analysed for DNA and RNA sequences in order to analyse the changes in microbial community structure and diversity throughout the pilot period.
In order to decrease the hydraulic retention time in future pilots as well as increase the organic loading rate of the pilot units, several potential modifications were noted during this pilot. These modifications will be further developed and implemented in future testing, which will be situated on the distillery site itself.
A collaboration between three Okinawan businesses supported the project by providing logistical assistance. Expertise in waste water handling and system installations was provided by Create-ES, a local waste water company who led the collaboration. Independent chemical analysis of the influent and effluent produced by the pilot was performed by The Okinawa Environmental Science Centre (OPESC). Technical expertise and marketing information was provided by the Okinawa Environmental Technology Centre (OPTEC).
A photo biological reactor was also tested alongside the MFC pilot, used to investigate the suitability of this type of microorganism to act as a tertiary treatment to the MFC system. It was shown that remaining nutrients within the effluent flow were reduced to a level suitable for surface water discharge of the final effluent. This technology will be further investigated as to its role to provide valuable by-products from produced biomass.

Figure 1: Overview picture of reactor and testing equipment setup.
3.2 A semi-automated genome annotation comparison and integration scheme and its application to the annotation of an electricity generating bacterium
Several genome annotation servises have been developed in recent years, such as IMG/ER from the Joint Genome Institute, the National Microbial Pathogen Data Resource's RAST server, JCVI annotation service and IGS annotation engine. These services can greatly reduce the cost and human efforts needed for annotating genome sequences. However, the functional annotation results from different services are often not the same. Furthermore, it is very difficult to compare the annotation results from different services due to the lack of standard IDs used in annotation. To address these problems we have developed a semi-automated scheme that is capable of comparing functional annotations from different sources and consequently obtaining a concensus genome functional annotation result. Our scheme is divided into two steps: annotation comparison and annotation determination. In the annotation comparison step, we employed gene synonym lists to tackle term difference problems. Multiple techniques from information retrieval such as Tokenization, Lemmatization were used to preprocess the functional annotations. The complete comparison processes between IGS and RAST is shown in Figure 2 using Arcobacter butzleri ED-1 (Arc-ED), a newly sequenced electrogenic bacteria of Epsilon-proteobacteria class as an example. Using our approach 87% of the annotations can be automatically compared, while only 39% when using exact ID/name matching.
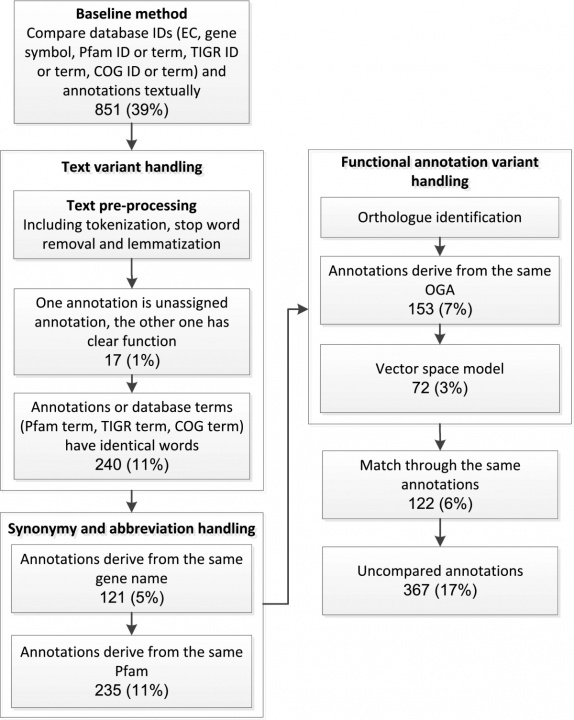
Figure 2: Flowchart of the gene annotation comparison procedure. The boxes represent the processes used to compare the annotations and the figures in each node the annotation comparison number and rate.
After obtaining the pair-wise comparison results between different annotation services, we constructed a decision tree to derive a consensus genome annotation result. Without any biological evidence, we could not assert which annotation was more reliable. Therefore we assumed that the gene annotations with the majority supports ought to be of high reliability. The work flow to obtain the consensus annotation is shown on Figure 3. To show the effectiveness of our approach, we applied our method to six published phylogenetically different genomes and using the literature-based E.coli annotation results from EcoCyc as a golden criteria to evaluate the reliability of the consensus annotation. The result showed that by integrating information from different resources, our scheme does produce better results than the individual services.
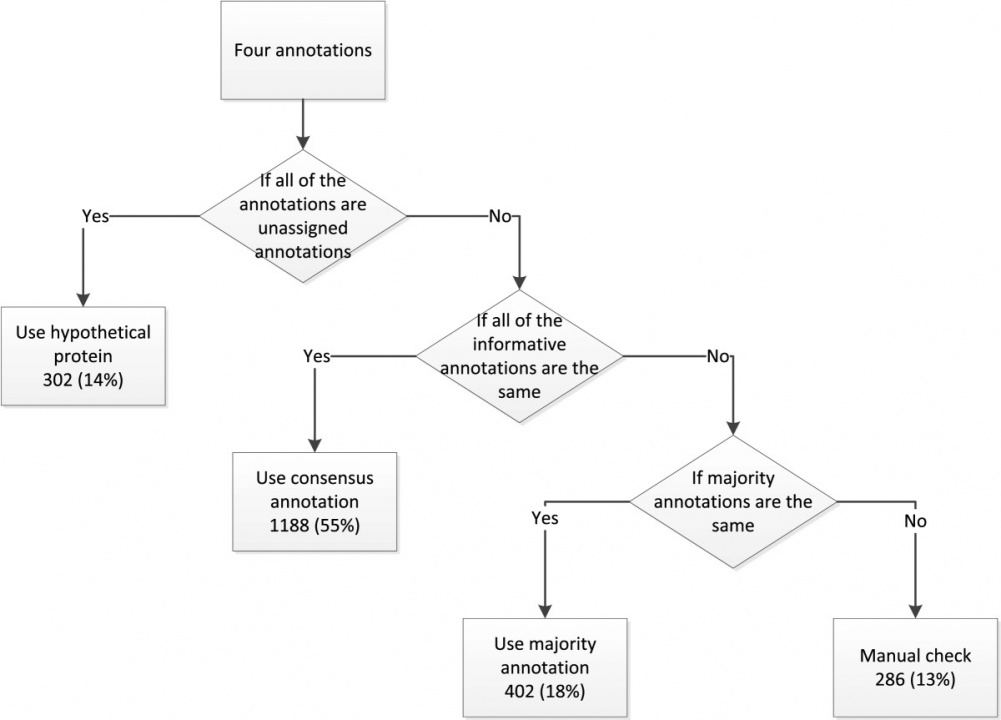
Figure 3: Flowchart of gene annotation determination procedure.
3.3 Regulation of human gamma-glutamyltransferase (GGT)
Recent findings show that GGT test has a predictive value as a sensitive biomarker of all-cause mortality risk. Despite the test is routinely has been used in clinics since 1960s for determination of liver tissue damage, the mechanisms behind the regulation of the measured serum human GGT level remains unclear. Aiming to uncover the mechanism of the measured GGT regulation and employing systems biology and bioinformatics tools we show that: 1) GGT assay measurements correspond to GGT1 protein, one of several proteins of the GGT protein family; 2) despite the importance of the GGT1 protein post-translational modification for the enzyme maturation, it is not the main mechanism of the enzyme activity regulation, and the focus should be on the regulation of its gene expression; 3) the list of TFs that participate in the GGT1 promoter regulation consist of confirmed TFs (for exampleSp1, c-Jun and ATF-2) and proposed TFs (for example p53, Nrf2 and PU.1); 4) signalling pathways that lead to the TFs in connection to GGT promoter are TNF-alpha receptor signalling pathways; 5) proposed new connection to various diseases and drugs. In addition, the result of our analysis allows us proposing a new mechanism of GGT regulation that differs from the regulation of other enzymes of the glutamyl cycle: toxin-induced mechanism of GGT enzyme up-regulation.
4. Publications
4.1 Journals
- Goryachev, A.B., Goryanin, I.I. (2012). Systems biology: desighn principles of whole biological systems. Preface. Advances in experimental medicine and biology 736, v
- De Ferrari, L., Aitken, S., van Hemert, J., Goryanin, I. (2012). EnzML: multi-label prediction of enzyme classes using InterPro signatures. BMC Bioinformatics 13 (1), 61.
- Sorokina, O., Goryanin, I. (2012). Theme issue modelling for pharmaceutical sciences. Preface. Europian Journal of Pharmaceutical Sciences 46 (4), 189.
- Adams, R., Clark, A., Yamaguchi, N., Hanlon, N., Tsorman, N., Ali, S., Lebedeva, G., Goltsov, A., Sorokin, A., Akman, O.E., Troein, C., Millar, A.J., Goryanin, I., Gilmore, S. (2013). SBSI: an extensible distributed software infrastructure for parameter estimation in systems biology. Bioinformatics 29 (5), 664-665.
- Thiele, I., Swainston, N., Fleming, R.M.T., Hoppe, A., Sahoo, S., Aurich, M., Haraldsdottir, H., Mo, M.L., Rolfsson, O., Stobbe, M.D., Thorleifsson, S.G., Agren, R., Bolling, C., Bordel, S., Chavali, A.K., Dobson, P., Dunn, W.B., Endler, L., Hala, D., Hucka, M., Hull, D., Jameson, D., Jamshidi, N., Jonsson, J.J., Juty, N., Keating, S., Nookaew, I., Le Novere, N., Malys, N., Mazein, A., Papin, J.A., Price, N.D., Selkov, E., Sigurdsson, M.I., Simeonidis, E., Sonnenschein, N., Smallbone, K., Sorokin, A., van Beek, J. HGM, Weichart, D., Goryanin, I., Nielsen, J., Westerhoff, H.V., Kell, D.B., Mendes, P., Palsson, B. (2013). A community-driven global reconstruction of human metabolism. Nature Biothechnology 31 (5), 419-425.
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Yomei high school students visit to OIST: "How I became a scientist" (August 29, 2012, OIST auditorium).
- Kyuyo high school students visit to OIST: "Where I am from and why I like science" (March 19, 2013, OIST auditorium).
- OIST science lesson for Iriomote and Ishigaki school students: "Part 1. Banana DNA extraction. Part 2. Assembling microbial fuel cell" (January 26, 2013, Iriomote ).
- Internal journal club on Development and evolution: "Cell typing by expression profile: A new look at an old problem" (July 12, 2012, OIST C016).
- External seminar in Moscow State University: "Microbial fuel cells" (December 27, 2012, MSU).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar
- Date: March 7, 2013
- Venue: OIST Campus Seminar Room C209
- Speaker: Kazuya Watanabe, Ph.D. (Professor, Tokyo University of Pharmacy and Life Science)
- Title: Microbial electric communications
6.2 Seminar
- Date: March 11, 2013
- Venue: OIST Campus Seminar Room B250
- Speaker: Jonathan Trent, Ph.D. (Senior Scientist at NASA Ames Research Center, Visiting Professor at Tokyo University of Agriculture and Technology and Adjunct Professor at the University of California, Santa Cruz)
- Title: OMEGA: For cultivating a sustainable future



