FY2019 Annual Report
G0 Cell Unit
Professor Mitsuhiro Yanagida

Abstract
The G0 Cell Unit studies how cells in the proliferative (dividing) or quiescent (non-dividing) phase respond to nutritional shifts (e.g., nitrogen source starvation and glucose limitation). We identify gene products (mostly proteins) and chemical factors (small molecules, metabolites) that affect adaptation mechanisms in order to understand the basis for longevity of long-term quiescent cells. We employ fission yeast as a eukaryotic cell model as the great majority of this organism’s genes are conserved in human. In addition, we are studying chromosomal regulatory mechanisms involving condensin, cohesin complexes (which lead to proper chromosome segregation in proliferating cells), and other nutrient adaptation-related nuclear chromatin proteins. This line of studies aims to understand dynamics of nuclear chromatin in response to nutritional cues. Study on human blood metabolomics initiated from comparative study with fission yeast metabolomics now directly aims to identify and understand the roles of metabolites intimately related to human aging.
Our current principal research projects may be summarized as below:
(1) Metabolomic approach to human aging and aging-related diseases.
(2) Understanding cell regulation in response to nitrogen deprivation.
(3) Understanding cell regulation in response to glucose starvation.
(4) Understanding chromosomal regulatory mechanisms involving condensin, cohesin complexes, and other nutrient adaptation-related nuclear proteins.
In FY2019, we published 5 original articles on research topics including chromosome regulatory mechanisms, genetic regulation for nutritional response, and human metabolomic analysis and filed 2 patents, as listed in 3. Activities and Findings, 4. Publications, and 5. Intellectual Property Rights and Other Specific Achievements.
1. Staff
- Dr. Takeshi Hayashi, Staff Scientist (until April)
- Dr. Susumu Morigasaki, Staff Scientist (until August)
- Dr. Norihiko Nakazawa, Staff Scientist (until July)
- Dr. Kenichi Sajiki, Staff Scientist (until August)
- Dr. Xingya Xu, Staff Scientist (from August)
- Dr. Haifeng Zhang, Postdoctoral Scholar (unitl April)
- Dr. Michiko Suma, Postdoctoral Scholar
- Dr. Yunfui Zheng, Postdoctoral Scholar (from January, 2020)
- Ms. Orie Arakawa, Technician
- Ms. Ayaka Mori, Technician
- Ms. Yuria Tahara, Technician
- Dr. Takayuki Teruya, Technician
- Ms. Risa Uehara, Technician
- Ms. Li Wang, Technician (from October)
- Ms. Chikako Sugiyama, Research Unit Administrator
2. Collaborations
- Theme: Analysis of human blood metabolites, involved in aging and aging-related diseases
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Takumi Mikawa, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Masahiro Kameda, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Comparative analysis of blood metabolome between healthy people and metabolic abnormality patients
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Masuzaki, Division of Endocrinology, Diabetes and Metabolism, Hematology, Rheumatology (Second Department of Internal Medicine), Graduate School of Medicine, University of the Ryukyus
- Theme: Blood metabolome analysis in abnormal cerebral function
- Type of collaboration: Joint research
- Researchers:
- Dr. Yasuhide Fukuji, Director, National Hospital Organization Ryukyu Hospital
- Dr. Taku Otsuru, Deputy Director, National Hospital Organization Ryukyu Hospital
3. Activities and Findings
3.1 Suppressor screening reveals common kleisin–hinge interaction in condensin and cohesin, but different modes of regulation
Cell divides to duplicate itself to form two daughter cells, that’s how all living things grow and survive. In each division, cell goes through a cell cycle, during which DNA (where the genetic information was stored) replicates. Sister DNAs are held together, before separation, by a protein complex called cohesin. Condensin is another protein complex that adapts a similar structure organization and play a fundamental role in sister DNA segregation. However, how cohesin and condensin complexes are formed, organized, and regulated to hold (and later release from) sister DNAs are still not well understood by scientists.
Cohesin complex was usually thought to form a ring-like structure, which hold sister DNAs inside the ring. Cut1 is an enzyme that functions as a pair of scissors, cuts cohesin when it’s time to release sister DNAs inside. However, in an earlier work of us published in Proceedings for the National Academy of Sciences (PNAS) in 2018, we found evidences that couldn’t fit to the ring model: cleavage of cohesin by Cut1 is not necessary when mutations in cohesin were introduced. Together with other evidence, such as tadpole-like structure of cohesin molecules under atomic force microscopy, we proposed a new model: ‘hold-and-release’ model. In this model cohesin is more like a jaw that holds chromatids in place and then opens to allow chromatin to move, without the need for Cut1 to break open the complex.
In a new study published in Proceedings for the National Academy of Sciences (PNAS) in 2019, we challenged to provide more evidences to support our ‘hold-and-release’ model, and find out how their functions are regulated by other pathways.
Isolation of extragenic suppressors is a powerful tool for identifying genes with protein products that function in the same process as a gene of interest, or that physically interact with the protein product of that gene. We have developed a powerful technology to identify suppressors easily using the cutting-edge next-generation sequencing. Based on our technologies, the study starts from genetic screening to identify genetic suppressors (second mutations) that can bypass the lethality caused by the first mutation located in cohesin or condensin subunits.
The problem we encountered is lack of temperature-sensitive (mutants that can grow at room temperature, but can’t grow at high temperature) or cold-sensitive (mutants that can grow at room temperature, but can’t grow at low temperature) mutants with mutations in cohesin or condensin subunits. To solve this problem, we developed an efficient method to isolate temperature-sensitive or cold-sensitive mutants with single amino acid substitutions targeted to the domain that is of interest. Please look at section 3.3 for the description of the technology.
Both cohesin and condensin contains two globular domains: a head domain and a hinge domain, that are connect by rope-like coiled-coils. In the ring model, the head domain and the hinge domain are far apart. However, in our ‘hold-and-release’ model, the head domain and the hinge domain are together and interact. We asked the question: If we perform suppressor screen using the temperature-sensitive or cold-sensitive mutants with mutation in hinge domain, can we get suppressor mutation in head domain? Ts or cs mutations, and suppressor mutations, always reflect physical interaction. Therefore, if we can identify hinge mutation suppressors in head domain, the result will support our model that hinge and head interact. Otherwise, it will support ring model that hinge and head are far apart.
Surprisingly, suppressors of hinge ts mutants were mapped in head domain, which support our ‘hold-and-release’ model very well. The results were true for both cohesin and condensin, therefore the ‘hold-and-release’ model applies to both cohesin and condensin. By analyzing the suppressor mutation localization, we found another amazing results that the mutations focus on a very narrow region in a domain called helix-turn-helix motif of kleisin that binds to head domain. So the helix-turn-helix motif of kleisin is very important for head-hinge interaction.
In addition to the findings above, we identified multiple suppressors, that are not rescuing physical interaction, but biological functions. Overall, although condensin and cohesin are similar in structural organization, they are regulated by distinct pathways: condensin is regulated by post-translational modifications (such as SUMOylation and phosphorylation), while cohesin is regulated by loading/releasing.
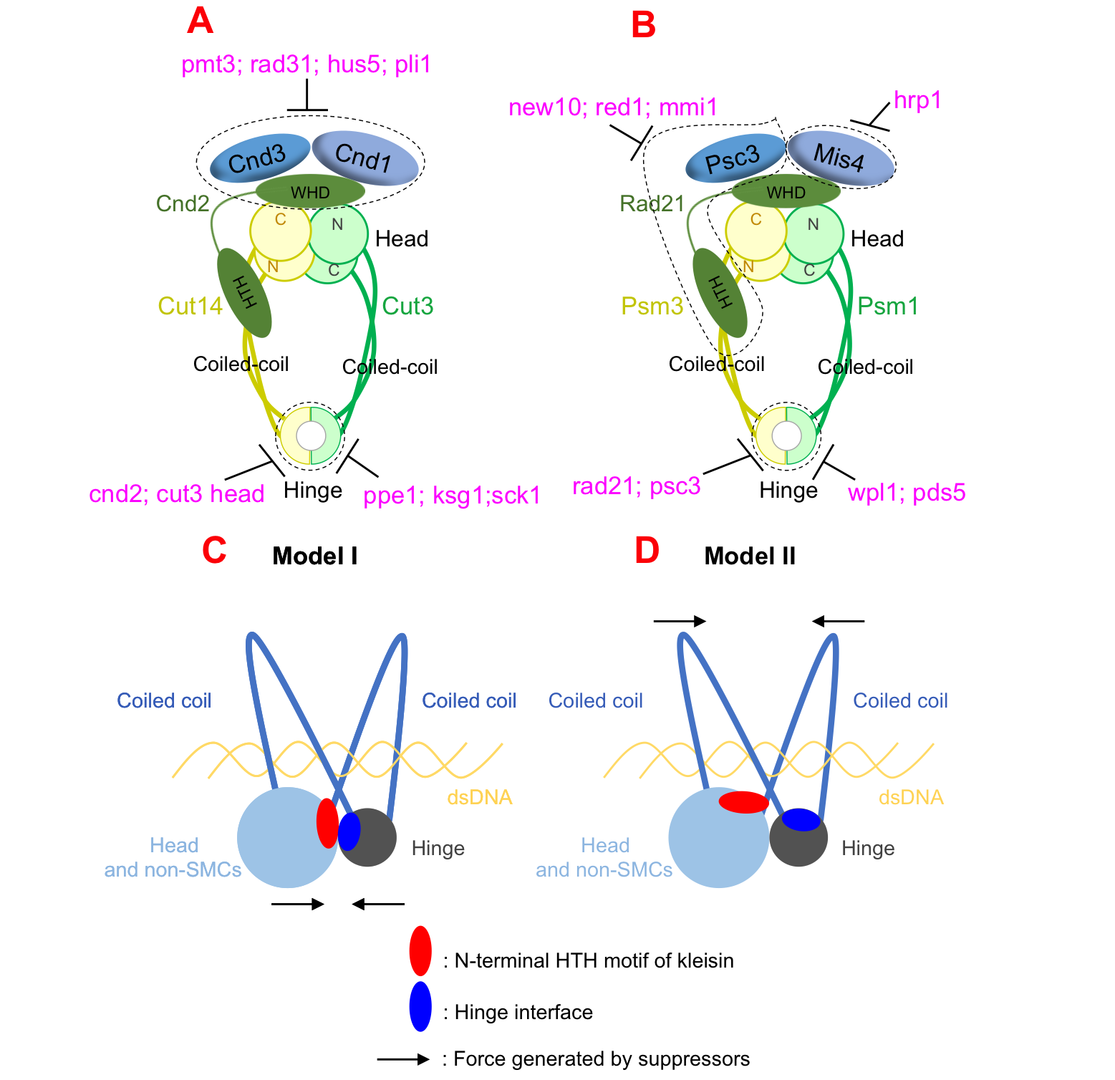
Figure 1: Condensin and cohesin are regulated differently, but they may adopt a similar organization in which hinge and head interact.
Although we have more and more evidences to support our ‘hold-and-release’ model, we have little knowledge about how DNA and ATP affect cohesin and condensin’s structures and activities. Current evidences indicates that cohesin and cohesin’s structures change after binding to DNA, and ATP plays a critical role in this structural re-organization. These questions will be helpful to answer how cohesin and condensin work dynamically in the future.
3.2 Condensin locates at transcriptional termination sites in mitosis, possibly releasing mitotic transcripts
Condensin is an essential component of chromosome dynamics, including mitotic chromosome condensation and segregation, DNA repair, and development. Genome-wide localization of condensin is known to correlate with transcriptional activity. The functional relationship between condensin accumulation and transcription sites remains unclear, however.
By constructing the auxin-inducible degron strain of condensin, herein we demonstrate that condensin does not affect transcription itself. Instead, RNA processing at transcriptional termination appears to define condensin accumulation sites during mitosis, in the fission yeast Schizosaccharomyces pombe. Combining the auxin-degron strain with the nda3 β-tubulin cold sensitive (cs) mutant enabled us to inactivate condensin in mitotically arrested cells, without releasing the cells into anaphase. Transcriptional activation and termination were not affected by condensin’s degron-mediated depletion, at heat-shock inducible genes or mitotically activated genes. On the other hand, condensin accumulation sites shifted approximately 500 bp downstream in the auxin-degron of 5’-3’ exoribonuclease Dhp1, in which transcripts became aberrantly elongated, suggesting that condensin accumulates at transcriptionally terminated DNA regions. Growth defects in mutant strains of 3’-processing ribonuclease and polyA cleavage factors were additive in condensin temperature-sensitive (ts) mutants.
Considering condensin’s in vitro activity to form double-stranded DNAs from unwound, single-stranded DNAs or DNA-RNA hybrids, condensin-mediated processing of mitotic transcripts at the 3'-end may be a prerequisite for faithful chromosome segregation.
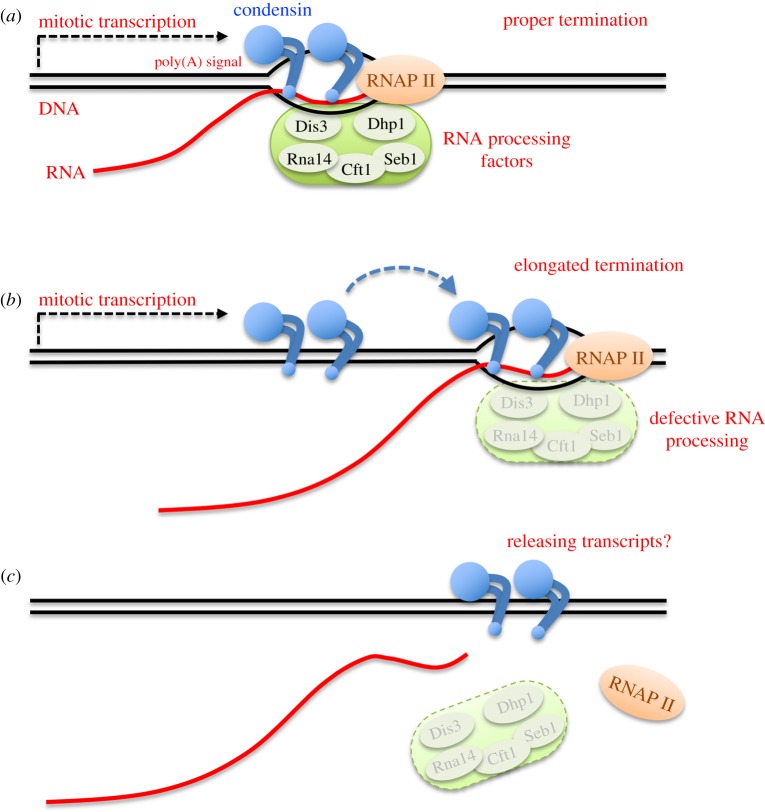
Figure 2: Possible action of condensin at transcriptional termination sites for releasing mitotic RNA transcripts.
3.3 Isolation of Fission Yeast Condensin Temperature-Sensitive Mutants with Single Amino Acid Substitutions Targeted to Hinge Domain
Essential genes cannot be deleted from the genome; therefore, temperature-sensitive (ts) mutants and cold-sensitive (cs) mutants are very useful to discover functions of essential genes in model organisms such as Schizosaccharomyces pombe and Saccharomyces cerevisiae.
To isolate ts/cs mutants for essential genes of interest, error-prone mutagenesis (or random mutagenesis) coupled with in vitro selection has been widely used. However, this method often introduces multiple silent mutations, in addition to the mutation responsible for ts/cs, with the result that one cannot discern which mutation is responsible for the ts/cs phenotype. In addition, the location of the responsible mutation introduced is random, whereas it is preferable to isolate ts/cs mutants with single amino acid substitutions, located in a targeted motif or domain of the protein of interest.
In our previous studies, ts or cs mutants with specific phenotypes were isolated by error-prone mutagenesis or forward genetic screening of the collection of ts/cs mutants. Many of them contain substitutions from Leucine (L)/Serine (S) to Proline (P) or from Glycine (G)/Alanine (A) to Aspartic Acid (D)/Glutamic Acid (E) (L/S -> P or G/A -> D/E), so L/S -> P or G/A -> D/E often caused ts or cs.
In this study, we have developed a method to isolate ts/cs mutants with single amino acid substitutions in targeted regions using site-directed mutagenesis. This method takes advantage of the empirical fact that single amino acid substitutions (L/S -> P or G/A -> E/D) often cause ts or cs. Application of the method to condensin and cohesin hinge domains was successful: ~20% of the selected single amino acid substitutions turned out to be ts or cs. This method is versatile in fission yeast and is expected to be broadly applicable to isolate ts/cs mutants with single amino acid substitutions in targeted regions of essential genes.
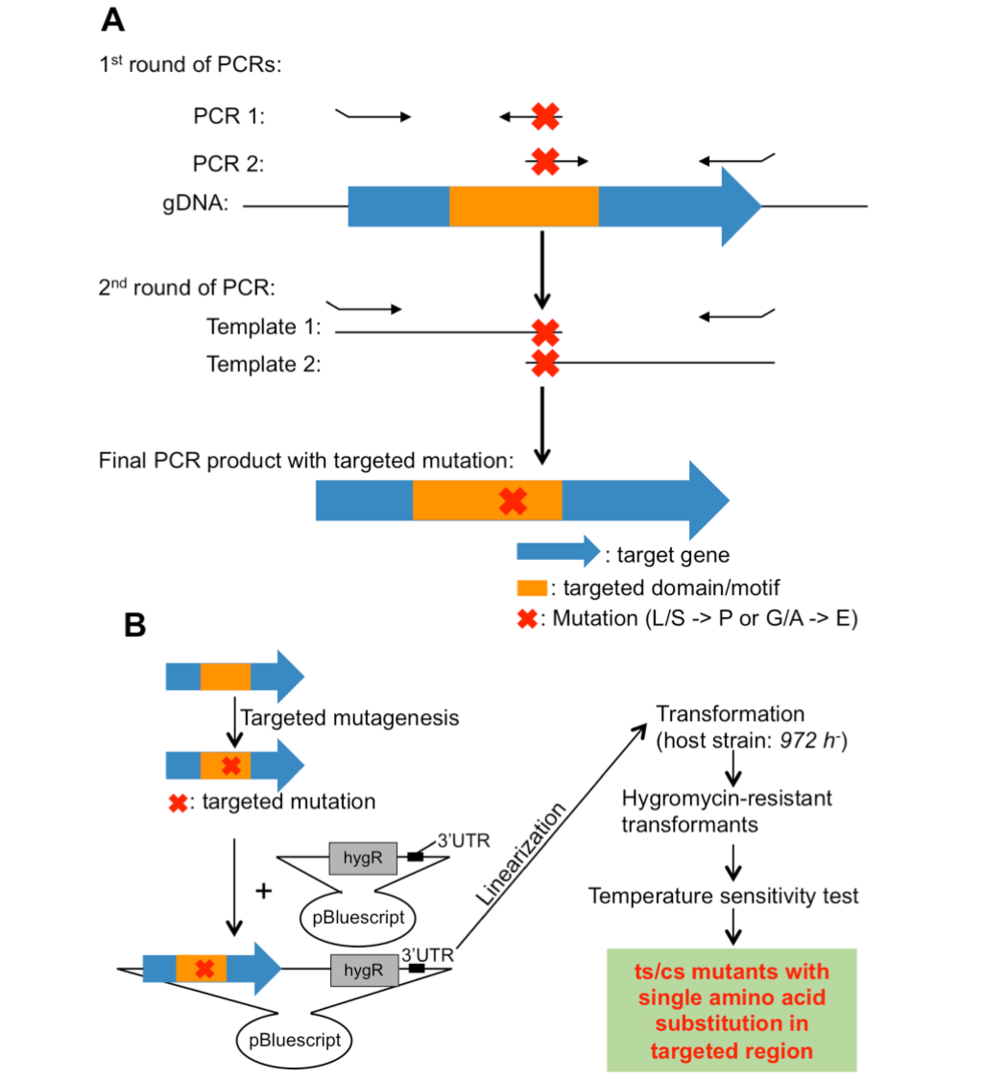
Figure 3: Site-directed mutagenesis. (A) Strategy to introduce targeted mutations into target genes. (B) Site-directed mutagenesis and isolation of ts/cs mutants.
3.4 Negative regulation of the Mis17-Mis6 centromere complex by mRNA decay pathway and EKC/KEOPS complex in Schizosaccharomyces pombe
A centromere is a part of a chromosome and it plays a crucial role in equal sister chromatid separation. Kinetochore is a protein complex assembled at each centromere, it associates with microtubules and motor proteins that segregate the divided sister chromatids toward the spindle poles in mitosis. Kinetochores in metaphase chromosomes must be bioriented toward opposite spindle poles to ensure correct sister chromatid separation (Rieder et al. 1995).
Deposition of the centromere-specific histone H3 variant, spCENP-A/Cnp1, is vital for the formation of centromere-specific chromatin and the Mis17-Mis6 complex of the fission yeast Schizosaccharomyces pombe is required for this deposition
In this study, we identified extragenic suppressors for a Mis17-Mis6 complex temperature-sensitive (ts) mutant, mis17-S353P, using whole-genome sequencing. The large and small daughter nuclei phenotype observed in mis17-S353P was greatly rescued by these suppressors. Suppressor mutations in two ribonuclease genes involved in the mRNA decay pathway, exo2 and pan2, may affect Mis17 protein level, as mis17 mutant protein level was recovered in mis17-S353P exo2 double mutant cells. Suppressor mutations in EKC/KEOPS complex genes may not regulate Mis17 protein level, but restored centromeric localization of spCENP-A/Cnp1, Mis6 and Mis15 in mis17-S353P. Therefore, the EKC/KEOPS complex may inhibit Mis17-Mis6 complex formation or centromeric localization. Mutational analysis in protein structure indicated that suppressor mutations in the EKC/KEOPS complex may interfere with its kinase activity or complex formation. Our results suggest that the mRNA decay pathway and the EKC/KEOPS complex negatively regulate Mis17-Mis6 complex-mediated centromere formation by distinct and unexpected mechanisms.
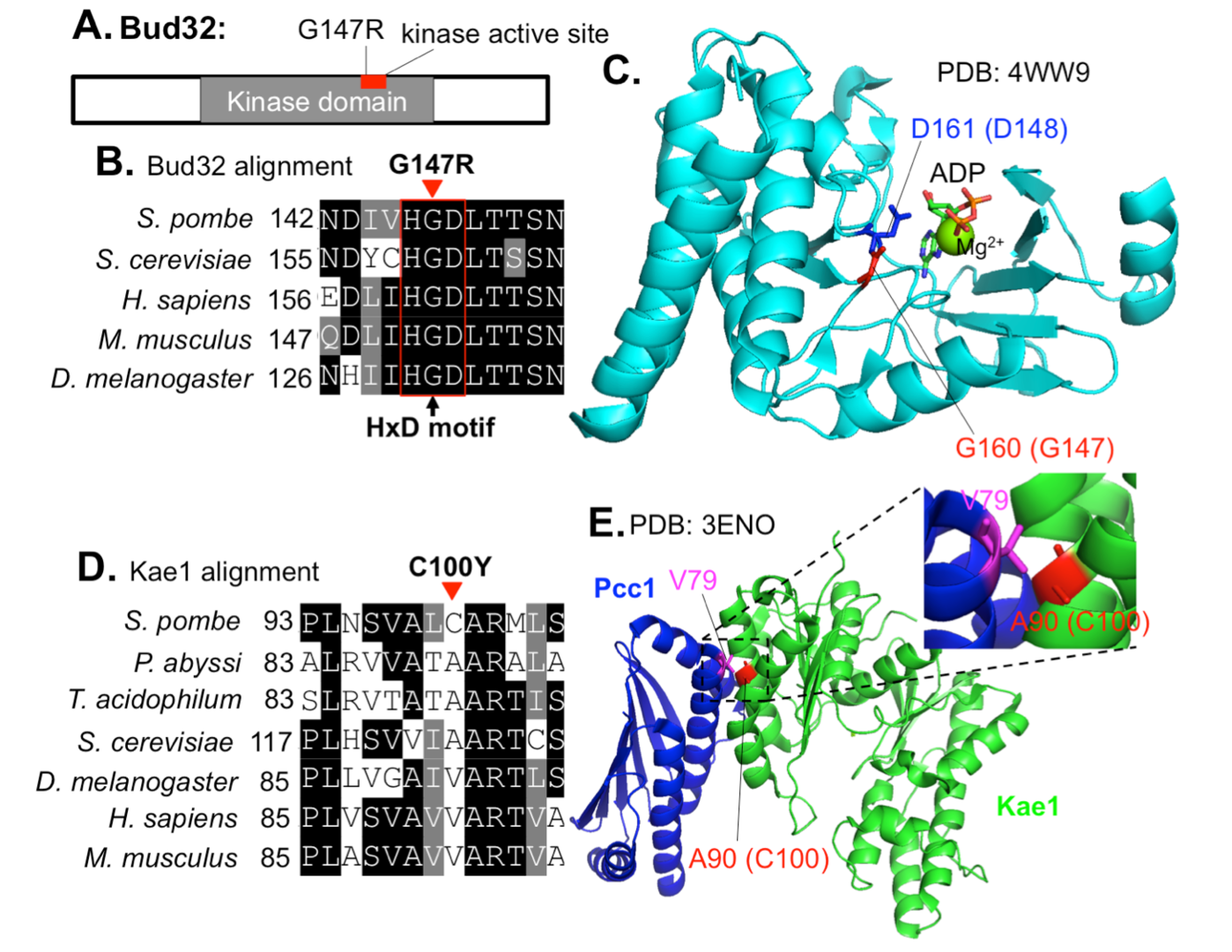
Figure 4: Localization of mis17-S353P suppressor mutations in EKC/KEOPS structures
3.5 Coordinated Roles of the Putative Ceramide-Conjugation Protein, Cwh43, and a Mn2+-Transporting, P-Type ATPase, Pmr1, in Fission Yeast
Genetically controlled mechanisms of cell division and quiescence are vital for responding to changes in the nutritional environment and for cell survival. Previously, we have characterized temperature-sensitive (ts) mutants of the cwh43 gene in fission yeast, Schizosaccharomyces pombe, which is required for both cell proliferation and nitrogen starvation-induced G0 quiescence. Cwh43 encodes an evolutionarily conserved transmembrane protein that localizes in endoplasmic reticulum (ER). Defects in this protein fail to divide in low glucose and lose mitotic competence under nitrogen starvation, and also affect lipid metabolism.
Here, we identified mutations of the pmr1 gene, which encodes an evolutionarily conserved Ca2+/Mn2+-transporting P-type ATPase, as potent extragenic suppressors of ts mutants of the cwh43 gene. Intriguingly, these pmr1 mutations specifically suppressed the ts phenotype of cwh43 mutants, among five P-type Ca2+- and/or Mn2+-ATPases reported in this organism. Cwh43 and Pmr1 co-localized in the ER. In cwh43 mutant cells, addition of excessive manganese to culture media enhanced the severe defect in cell morphology, and caused abnormal accumulation of a cell wall component, 1, 3-β-glucan. In contrast, these abnormal phenotypes were abolished by deletion of the pmr1+ gene, as well as by removal of Mn2+ from the culture medium. Furthermore, nutrition-related phenotypes of cwh43 mutant cells were rescued in the absence of Pmr1. Our findings indicate that the cellular processes regulated by Cwh43 are appropriately balanced with Pmr1-mediated Mn2+ transport into the ER.
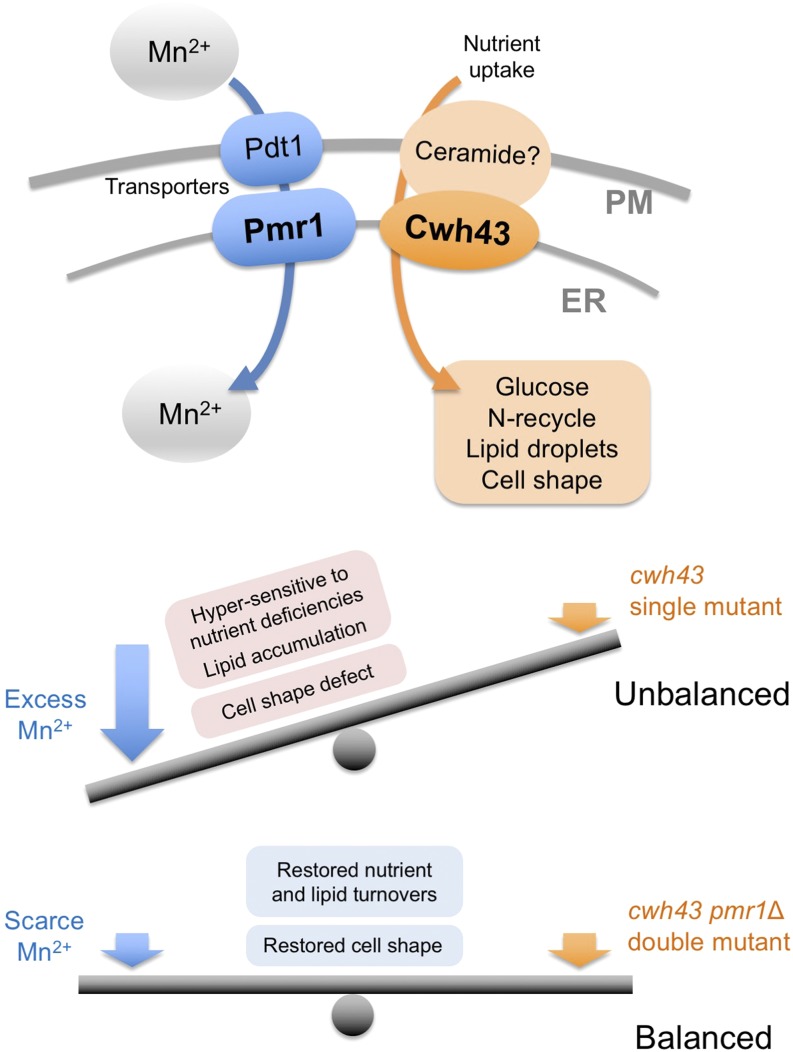
Figure 5: Balanced action between Cwh43 and Pmr1-mediated Mn2+ transport.
4. Publications
4.1 Journals
- Nakazawa N, Arakawa O, Yanagida M (2019) Condensin locates at transcriptional termination sites in mitosis, possibly releasing mitotic transcripts. Open Biol. [PubMed]
- Nakazawa N, Xu X, Arakawa O, Yanagida M (2019) Coordinated Roles of the Putative Ceramide-Conjugation Protein, Cwh43, and a Mn2+-Transporting, P-Type ATPase, Pmr1, in Fission Yeast. G3 (Bethesda) [PubMed]
- Nakazawa, N (2019) Physiological role of “Ceramide remodelase” revealed by two studies in fission and budding yeasts. KAGAKU TO SEIBUTSU 57, 334-336, doi: 10.1271/kagakutoseibutsu.57.334
- Xu X, Yanagida M (2019) Suppressor screening reveals common kleisin-hinge interaction in condensin and cohesin, but different modes of regulation. Pcoc Natl Acad Sci USA (PNAS) [PubMed]
- Xu X, Nakazawa N, Wang L, Arakawa O, Yanagida M (2019) Negative Regulation of the Mis17-Mis6 Centromere Complex by mRNA Decay Pathway and EKC/KEOPS Complex in Schizosaccharomyces pombe. G3 (Bethesda) [PubMed]
- Xu X, Yanagida M (2019) Isolation of Fission Yeast Condensin Temperature-Sensitive Mutants with Single Amino Acid Substitutions Targeted to Hinge Domain. G3 (Bethesda) [PubMed]
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Teruya, T. A Commercialization verification of a service that comprehensively monitors physiological functions using metabolites as an index, Program of Start-up incubation from COre REsearch (SCORE) Demo Day, JST, Tokyo, Japan, 2020.2.1
- Yanagida, M. Hinge-head interaction in Condensin and Cohesin, The 37th Chromesome Workshop, Niigata, Japan, 2019.12.22-24
- Xu, X. Hold-and-release model of Condensin and Cohesin, The 37th Chromesome Workshop, Niigata, Japan, 2019.12.22-24
- Sajiki, K. Kinase and phosphatases for quiescent survival, Cold Spring Harbor Asia Conference: Kinase & Phosphatase Signaling, Suzhou, China, 2019.12.9-13
- Xu, X. Suppressor screening reveals common organization of condensin and cohesin, 2019 Cold Spring Harbor meeting: Yeast Reseach; Origins, Insights, Breakthroughs, Cold Spring Harbor, New York, USA, 2019.10.23-26.
- Sajiki, K. Gene network essential for G0 phase, Konan University, Japan, 2019.6.13
- Teruya, T. Technology for aging degree determination based on metabolite information of human blood, urine, and saliva, New Technology Presentation Meeting, JST, Tokyo, Japan, 2019. 6.11
5. Intellectual Property Rights and Other Specific Achievements
- Yanagida M, Kondoh H, Kameda M, Teruya T. Novel indicators of frailty include blood metabolites for antioxidation, cognition, and mobility. Provisional Japanese patent application, JP2019-182716 (filing date 03.10.2019)
- Yanagida M, Teruya T. METHOD FOR EVALUATING RISK OF TYPE 2 DIABETES USING BLOOD METABOLITES AS AN INDEX. Patent application, PCT/JP2019/44926 (filing date 15.11.2019)
6. Meetings and Events
Nothing to report
7. Other
Nothing to report.



