FY2017 Annual Report
G0 Cell Unit
Professor Mitsuhiro Yanagida

Abstract
The G0 Cell Unit studies how cells in the proliferative (dividing) or quiescent (non-dividing) phase respond to nutritional shifts (e.g., nitrogen source starvation and glucose limitation). We identify gene products (mostly proteins) and chemical factors (small molecules, metabolites) that affect adaptation mechanisms in order to understand the basis for longevity of long-term quiescent cells. We employ fission yeast as a eukaryotic cell model as the great majority of this organism’s genes are conserved in human. In addition, we are studying chromosomal regulatory mechanisms involving condensin, cohesin complexes (which lead to proper chromosome segregation in proliferating cells), and other nutrient adaptation-related nuclear chromatin proteins. This line of studies aims to understand dynamics of nuclear chromatin in response to nutritional cues. Study on human blood metabolomics initiated from comparative study with fission yeast metabolomics now directly aims to identify and understand the roles of metabolites intimately related to human aging.
Our current principal research projects may be summarized as below:
(1a) Methodological development for metabolomic analysis.
(1b) Comprehensive and quantitative human blood metabolomics.
(2) Understanding cell regulation in response to nitrogen deprivation.
(3) Understanding cell regulation in response to glucose starvation.
(4) Understanding chromosomal regulatory mechanisms involving condensin, cohesin complexes, and other nutrient adaptation-related nuclear proteins.
In FY2017, we published original articles on pathways that compensate for choromosome segregation defects identified by whole-genome sequencing of suppressor DNA mixtures, and on genetic defects rescued by rapamycin, as listed in 3. Activities and Findings and 4. Publications.
1. Staff
- Dr. Yung-Ju Chen, Researcher
- Dr. Takeshi Hayashi, Researcher
- Dr. Susumu Morigasaki, Researcher (from October)
- Dr. Norihiko Nakazawa, Researcher
- Dr. Paul-Emile Poleni, Researcher (until July)
- Dr. Kenichi Sajiki, Researcher
- Dr. Xingya Xu, Researcher
- Dr. Haifeng Zhang, Researcher
- Ms. Orie Arakawa, Technician
- Ms. Ayaka Mori, Technician
- Ms. Michiko Suma, Technician
- Ms. Yuria Tahara, Technician
- Ms. Junko Takada, Technician
- Dr. Takayuki Teruya, Technician
- Ms. Risa Uehara, Technician
- Ms. Li Wang, Technical staff
- Ms. Caroline Starzynski, OIST Student
- Mr. Hunter Barbee, OIST Student (from September to December)
- Mr. Haruhisa Goga, Visiting Researcher (from April to September)
- Ms. Chikako Sugiyama, Research Unit administrator
2. Collaborations
- Theme: Screening for low-glucose sensitive mutants in fission yeast
- Type of collaboration: Joint research
- Researchers:
- Professor Shigeaki Saitoh, Institute of Life Science, Kurume University
- Theme: Elucidation of adaptation mechanism to low glucose condition in fission yeast
- Type of collaboration: Joint research
- Researchers:
- Professor Kunihiro Ohta, Department of Life Science, Graduate School of Arts and Science, The University of Tokyo
- Theme: Analysis of human blood metabolites, involved in aging and aging-related diseases
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Takumi Mikawa, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Structural understanding of the mechanisms of chromosomal cohesion by cohesin and chromosomal condensation/segregation by condensin
- Type of collaboration: Joint research
- Researchers:
- Professor Chikashi Toyoshima, Institute of Molecular and Cellular Biosciences, the University of Tokyo
- Dr. Ryuta Kanai, Institute of Molecular and Cellular Biosciences, the University of Tokyo
- Theme: Imaging of molecular structures of condensin and cohesin complexes by the atomic force microscopy
- Type of collaboration: Joint research
- Researchers:
- Dr. Shige H. Yoshimura, Graduate Schoolo of Biostudies, Kyoto University
- Theme: Comparative analysis of blood metabolome between healthy people and metabolic abnormality patients
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Masuzaki, Division of Endocrinology, Diabetes and Metabolism, Hematology, Rheumatology (Second Department of Internal Medicine), Graduate School of Medicine, University of the Ryukyus
- Theme: Blood metabolome analysis in abnormal cerebral function
- Type of collaboration: Joint research
- Researchers:
- Dr. Yasuhide Fukuji, Director, National Hospital Organization Ryukyu Hospital
- Dr. Taku Otsuru, Deputy Director, National Hospital Organization Ryukyu Hospital
3. Activities and Findings
3.1 Whole-Genome Sequencing of Suppressor DNA Mixtures Identifies Pathways That Compensate for Chromosome Segregation Defects in Schizosaccharomyces pombe.
Suppressor screening is a powerful method to identify genes that, when mutated, rescue the temperature sensitivity of the original mutation. Previously, however, identification of suppressor mutations has been technically difficult. Due to the small genome size of Schizosaccharomyces pombe, we developed a spontaneous suppressor screening technique, followed by a cost-effective sequencing method. Genomic DNAs of 10 revertants that survived at the restrictive temperature of the original temperature sensitive (ts) mutant were mixed together as one sample before constructing a library for sequencing. Responsible suppressor mutations were identified bioinformatically based on allele frequency. Then, we isolated a large number of spontaneous extragenic suppressors for three ts mutants that exhibited defects in chromosome segregation at their restrictive temperature.
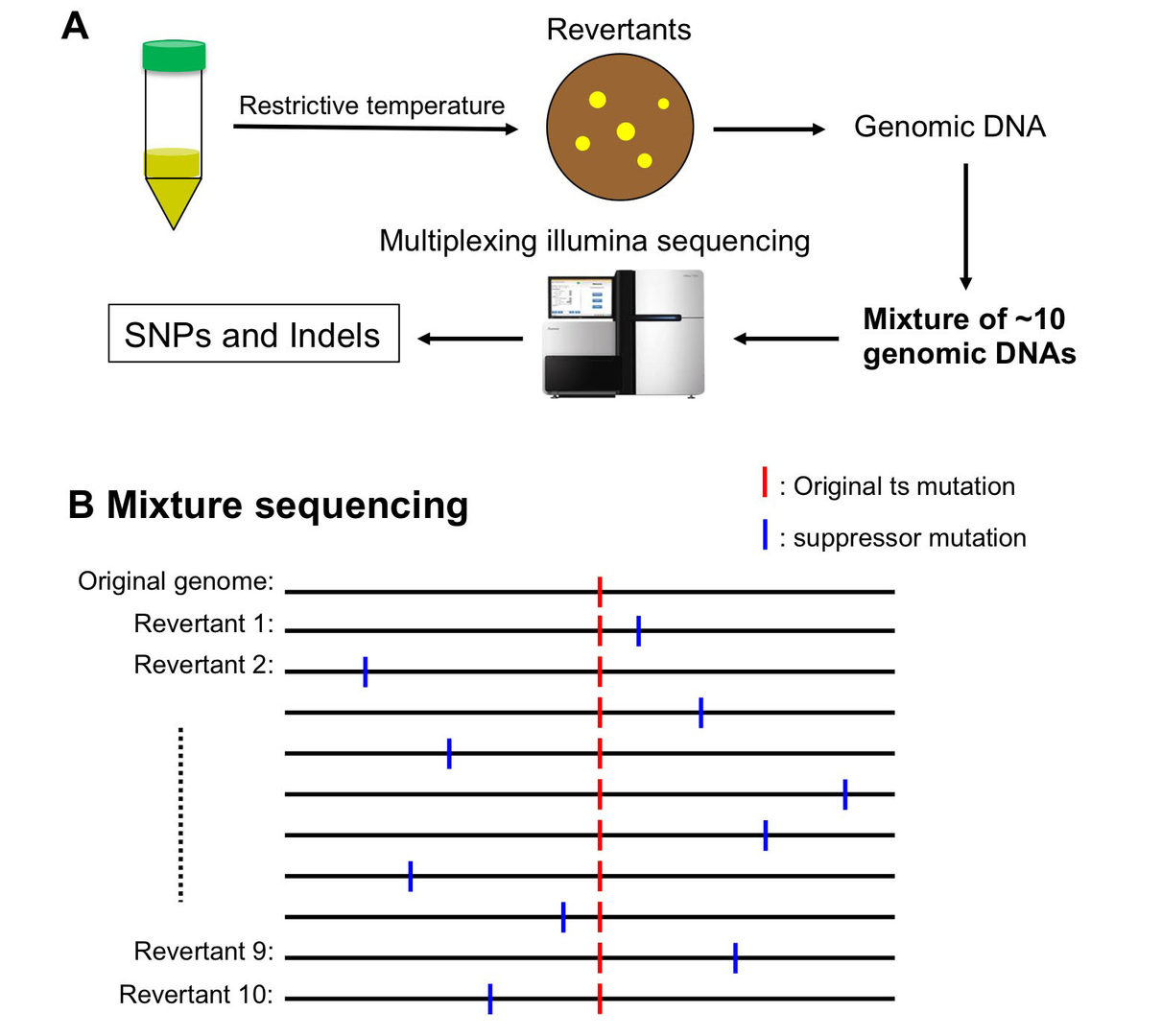
Screening provided new insight into mechanisms of chromosome segregation: loss of Ufd2 E4 multi-ubiquitination activity suppresses defects of an AAA ATPase, Cdc48. Loss of Wpl1, a releaser of cohesin, compensates for the Eso1 mutation, which may destabilize sister chromatid cohesion. The segregation defect of a ts histone H2B mutant is rescued if it fails to be deubiquitinated by the SAGA complex, because H2B is stabilized by monoubiquitination.
3.2 Genetic defects in SAPK signalling, chromatin regulation, vesicle transport and CoA-related lipid metabolism are rescued by rapamycin in fission yeast (From OIST news center article "Cure for Fission Yeast Genes Could Have Bigger Things Ahead" by Andrew Scott)
Scientists at the Okinawa Institute of Science and Technology Graduate University (OIST) have taken one step closer towards potential cures for several human genetic diseases, and the answers have been found in the humble cells of fission yeast.
Dr. Kenichi Sajiki and the team in Prof. Mitsuhiro Yanagida’s G0 Cell Unit at OIST have uncovered that rapamycin, a biological compound produced by bacteria, is able to “rescue” cells with genetic defects and maintain normal cell function. Their work was published in Open Biology.
Rapamycin is already in common use throughout the medical world as an immunosuppressant – stopping the function of the immune system. As such it is commonly prescribed to patients undergoing kidney transplants to prevent rejection of the new organs. It is also routinely used for cancer treatments, as well as for coating coronary stents. Rapamycin has also generated keen attention due to experiments that showed extended lifespan in mouse cells.
The OIST team has uncovered a new side to rapamycin, paving the way to new medical therapies for genetic diseases.
“This is the first systematic screen to find mutants that can be ‘cured’ by the drug. Previously we knew yeast chromosome diseases were cured by the same drug. Now Sajiki and more than ten authors discovered 12 more yeast gene diseases curable by the same drug,” says Prof. Yanagida.
Rapamycin inhibits cell growth and division – known as proliferation - by regulating the function of TOR Kinase, a main signaling enzyme that conveys information to the cell about nutritional conditions around it. The team hypothesized that if rapamycin could restore normal proliferation in cells with mutated DNA, it was possible that the affected genes conserved in humans responsible for genetic diseases could potentially be treated by rapamycin.
The team devised an experiment using fission yeast cells, each of which had a specific mutation in their DNA that created cell division defects when exposed to raised temperatures of 36°C. Testing a library of 1014 mutant strains of yeast. 45 of the mutants were “rescued” by rapamycin addition and began to divide normally once again.
Analyzing the genetic makeup of the rescued yeast cells, the lab team identified 12 different genes that were responsible for the temperature-triggered defects. With the source identified, the team now knew where to look for the answer to rapamycin’s restorative properties.
The genes are involved with four cellular function groups. One controls stress response signaling by creating a type of enzyme called Stress Activated Protein Kinase (SAPK). Left alone, mutated versions of these genes created SAPKs that were responsible for the malfunctions in experimental yeast cells, again causing abnormal cell growth and division.
Adding rapamycin saw these cells function as normal. The scientists observed that rapamycin essentially overrides the aberrant behavior of SAPK mutation so that resulting biochemical production is maintained at normal levels. This controls a delicate balance between SAPK and TOR Kinase that is key to keeping cells growing and dividing normally. Further groups of genes affected by rapamycin in this way include those responsible for cellular functions such as the exchange of chemicals between cells and the regulation of the genetic compound chromatin’s structure.
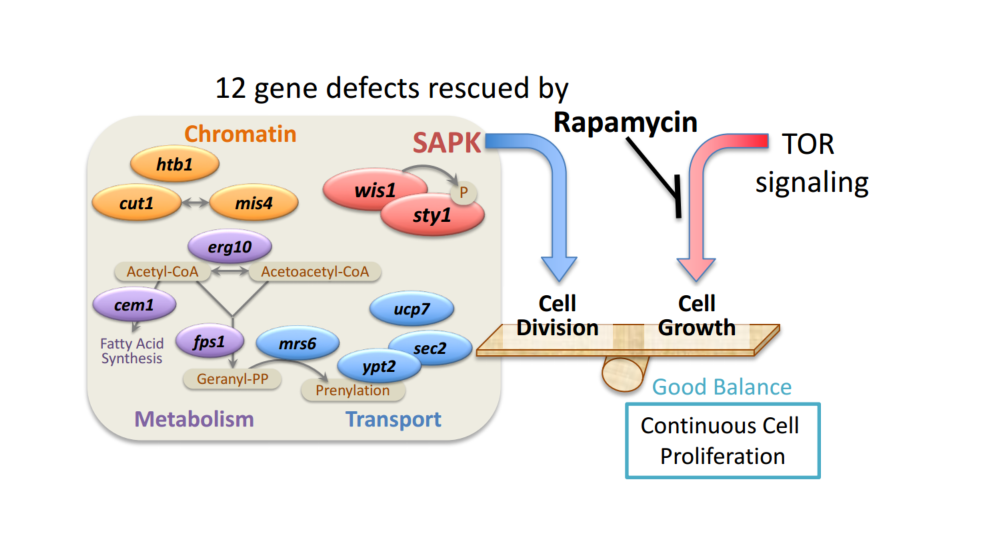
The study is a first step in opening new avenues of research, which could lead to cures for debilitating genetic conditions like Cornelia de Lange syndrome and cognitive development disorders.
Their identification of the method through which rapamycin works adds a critical new dimension to this already highly useful chemical: “We hope this research will give a new insight into how we can apply rapamycin in the future,” says Dr. Sajiki.
4. Publications
4.1 Journals
- Inoue H, Sugimoto S, Takeshita Y, Takeuchi M, Hatanaka M, Nagao K, Hayashi T, Kokubu A, Yanagida M, Kanoh J. CK2 phospho-independent assembly of the Tel2-associated stress-signaling complexes in Schizosaccharomyces pombe. Genes Cells. 22(1):59-70. (2017)
- Xu X, Wang L, Yanagida M. Whole-Genome Sequencing of Suppressor DNA Mixtures Identifies Pathways That Compensate for Chromosome Segregation Defects in Schizosaccharomyces pombe. G3 (Genes, Genomes, Genetics). 8(3):1031-1038(2018)
- Sajiki K, Tahara Y, Villar-Briones A, Pluskal T, Teruya T, Mori A, Hatanaka M, Ebe M, Nakamura T, Aoki K, Nakaseko Y, Yanagida M. Genetic defects in SAPK signalling, chromatin regulation, vesicle transport and CoA-related lipid metabolism are rescued by rapamycin in fission yeast. Open Biology. 8: 170261 (2018)
4.2 Books and other one-time publications
- Starzynski C. Investigating an evolutionary conserved metabolic mechanism contributing to G0 quiescence survival in S. pombe. Dissertation, 28 December (2017)
4.3 Oral and Poster Presentations
- Yanagida, M. Networks in separase mutant suppressors and human age-related metabolites, Genetic Networks Symposium, Canada Embassy, Tokyo, Japan, 27 April (2017).
- Yanagida, M. What Will Comparison of Fission Yeast and Human Blood Comprehensive Metabolomics Tell Us?, Pombe 2017, Banff, Canada, 14-19 May (2017).
- Hayashi, T. Fission yeast S-adenosylmethionine synthetase (Sam1) is required for cell growth and quiescence in coordination with TOR-related kinase, Pombe 2017, Banff, Canada, 14-19 May (2017)
- Nakazawa, N. Role of Cwh43, ceramide remodeling factor for GPI-anchored proteins, in nutrient utilization and lipid metabolism, Pombe 2017, Banff, Canada, 14-19 May (2017)
- Suma, M. Cnp3(CENP-C) prevents localization of Cnp1(CENP-A) to non-centromeric regions, Pombe 2017, Banff, Canada, 14-19 May (2017)
- Yanagida, M. Cohesin Clip Model Disclosed by Atomic Structure Assignment of Numerous Genetic Suppressors, Pombe 2017, Banff, Canada, 15 May (2017)
- Yanagida M. Cohesin clip model revealed by atomic structure assignment of genetic suppressors, UC Berkeley, USA, 18 May (2017)
- Yanagida M. Want to know about metabolites of old people, Gakushuin Univ., Tokyo, Japan, 10 June (2017)
- Yanagida M. Cohesin clip model, SMC Proteins 2017, Yamagata, Japan, 14 June (2017)
- Yanagida M. Can human aging be ‘molecularly’ measured?, Tsukuba Univ. Ibaragi, Japan, 22 June (2017)
- Yanagida M. Quantitative metabolomics analysis of human aging and fasting, Tohoku Forum for Creativity Thematic Program 2017 International Symposium 'Food Safety and Functional Evaluation', Tohoku Univ., Sendai, Japan, 7 August (2017)
- Nakazawa N. Ceramide remodelase Cwh43 maintains chronological lifespan and restrains excessive storage of neutral lipids in fission yeast, 50th Yeast Genetics and Molecular Biology Forum, Univ. of Tokyo, Japan, 13 September (2017)
- Sajiki K. SHK and GZE - Two gene groups required for the G0 phase entry regulation, 50th Yeast Genetics and Molecular Biology Forum, Univ. of Tokyo, Japan, 13 September (2017)
- Starzynski C. A conserved metabolic mechanism contributing to quiescence survival in S. pombe, 50th Yeast Genetics and Molecular Biology Forum, Univ. of Tokyo, Japan, 11 September (2017)
- Yanagida M. Pleiotropic phenotype in fission yeast mutants defective in the synthesis of the methyl donor S-adenosylmethionine, 50th Yeast Genetics and Molecular Biology Forum, Univ. of Tokyo, Japan, 11 September (2017)
- Nakazawa N. Ceramide remodelase Cwh43 maintains chronological lifespan and restrains excessive storage of neutral lipids in fission yeast, 89th Symposium for Genetics Society of Japan, Okayama Univ., Japan, 15 September (2017)
- Yanagida M. Metabolite linked with aging, The 38th Japan Society for Biomedical Gerontology Symposium, Kyoto, Japan, 14 October (2017)
- Yanagida M. Human aging-related metabolites and control of cellular quiescence, National Univ. of Singapore, 20 November (2017)
- Yanagida M. A Japanese scientist assesses the past 50 years and the future of life sciences and biotechnology, National Univ. of Singapore, 21 November (2017)
- Yanagida M. Human aging-related metabolites and control of cellular quiescence, Duke NUS Medical School, Singapore, 23 November (2017)
- Yanagida M. A Japanese scientist assesses the past 50 years and the future of life sciences and biotechnology, Duke NUS Medical School, Singapore, 24 November (2017)
- Yanagida M. Blood metabolites as molecular measures of human aging, Consoritum of Biological Sciences 2017, Kobe, Japan, 7 December (2017)
- Nakazawa N. Ceramide remodelase, Cwh43, controls proper responses to nitrogen and glucose, and maintains neutral lipid homeostasis in fission yeast, The 2018 annual meeting of the Japan Society of Bioscience, Biotechnology, and Agrochemistry, Meijo Univ., Nagoya, Japan, 17 March (2018)
- Yanagida M. Metabolites commonly present in human blood and fission yeast cell, The 2018 annual meeting of the Japan Society of Bioscience, Biotechnology, and Agrochemistry, Meijo Univ., Nagoya, Japan, 18 March (2018)
5. Intellectual Property Rights and Other Specific Achievements
5.1 Intellectual Property Rights
- Yanagida M, Kondoh H, Teruya T. A METHOD, AN APPARATUS, A SYSTEM AND A KIT FOR DETERMINING THE EXTENT OF AGING. Patent application, PCT/JP2017/009758, WO/2017/155100 (filing data 10.03.2017, publication data 14.09.2017).
6. Meetings and Events
6.1 Seminar
1. Title: Life-span regulation by environmental stresses in C. elegants
- Date: April 6, 2017
- Venue: C016, OIST Campus Lab1
- Speaker: Prof. Eisuke Nishida (Department of Cell and Developmental Biology, Graduate School of Biostudies, Kyoto University)
2. Title: Sensing mechanism of plasma membrane lipid asymmetry and cellular response
- Date: March 1, 2018
- Venue: D015, OIST Campus Lab1
- Speaker: Dr. Keisuke Obara (Assistant Professor, Graduate School of Science, Nagoya University)
7. Other
Nothing to report.



