FY2013 Annual Report
G0 Cell Unit
Professor Mitsuhiro Yanagida

Abstract
The regulation of cellular proliferation and quiescence (G0 phase) in response to environmental changes, such as the availability of nutrients, is a central issue in biology and medicine. Our research goal is to elucidate the molecular mechanisms that regulate pivotal events in cellular proliferation and quiescence, such as chromosome segregation, coupling of cell growth and division, and cellular adaptation to nutrient paucity. To study the molecular mechanisms in detail, we have adopted the fission yeast Schizosaccharomyces pombe, a simple unicellular eukaryote, as a model organism, and use post-genomic methods in combination with genetic approaches. In FY2013, we further evaluated the G0 phase induced by nitrogen starvation and the adaptation of S. pombe to glucose-limited conditions. We reported the results of our metabolomic analysis at the onset of nitrogen starvation in Metabolites (Sajiki et al., 2013), and the requirement for Klf1, a C2H2 zinc finger-transcription factor, in cell wall maintenance during long-term quiescence in PLOS ONE (Shimanuki et al., 2013). In addition, we investigated the biosynthesis of G0-related metabolites and various regulatory mechanisms of cellular proliferation. We also studied the health benefits of Okinawan produce and the background of Okinawan longevity as one of the R&D cluster projects in Okinawa, and reported the methods of metabolomic analysis of human blood, plasma, and red blood cells in comparison with S. pombe in Rinsho Ketsueki (Chaleckis et al., 2013).
1. Staff
- Dr. Takeshi Hayashi, Group leader
- Dr. Kazuki Kumada, Group leader
- Dr. Kenichi Sajiki, Researcher
- Dr. Norihiko Nakazawa, Researcher
- Dr. Takayuki Teruya, Researcher
- Dr. Xiaodong He, Researcher
- Dr. Emily Tsang, Researcher
- Dr. Rajesh Mehrotra, Researcher (from July, until December)
- Ms. Ayaka Mori, Technical staff
- Mr. Tomas Pluskal, Technical staff
- Ms. Yuria Tahara, Technical staff
- Mr. Masahiro Ebe, Technical staff
- Ms. Orie Arakawa, Technical staff
- Ms. Li Wang, Technical staff (Part timer, from May)
- Ms. Chikako Sugiyama, Research administrator
- Ms. Caroline Starzynski, OIST Student (from September)
- Mr. Xingya Xu, Special Research Student
- Mr. Raphael Chow, Research Intern (from June, until August)
2. Collaborations
- Theme: Metabolite structure determination using NMR
- Type of collaboration: Joint research
- Researchers:
- Associate Professor, Masaru Ueno, Department of Molecular Biotechnology, Graduate school of Advanced Science of Matter, Hiroshima University
- Theme: Identification of fission yeast mutants, which show growth defect under low glucose condition
- Type of collaboration: Joint research
- Researchers:
- Professor, Shigeaki Saitoh, Institute of Life Science, Kurume University
- Theme: Identification of the molecular mechanism, required for the maintenance of cell viability in low glucose condition
- Type of collaboration: Joint research
- Researchers:
- Professor, Kunihiro Ohta, Department of Life Science, Graduate School of Arts and Science, University of Tokyo
- Theme: Metabolomic analysis of human blood
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Takumi Mikawa, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Functional analysis of condensin complex in the regulation of chromosome segregation and gene expression
- Type of collaboration: Joint research
- Researchers:
- Dr. Takashi Sutani, Institute of Molecular Cellular Biosciences, The University of Tokyo
- Theme: Physiological/genetic analysis of adaptation process to low glucose environment in fission yeast
- Type of collaboration: Joint research
- Researchers:
- Dr. Kojiro Takeda, Department of Biology, Faculty of Science and Engineering, Konan University
- Theme: Metabolic analysis of active ingredients of health foods
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Metabolic analysis of active ingredients of health foods
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Masuzaki, Division of Endocrinology, Diabetes and Metabolism, Hematology, Rheumatology (Second Department of Internal Medicine), Graduate School of Medicine, University of the Ryukyus
- Theme: Dolphin blood metabolome analysis
- Type of collaboration: Joint research
- Researchers:
- Dr. Tadashi Maruyama, Marine Biodiversity Research Program, Japan Marine Science and Technology Center (JAMSTC)
- Mr. Yoshiaki Kamei, Okinawa Churaumi Aquarium, Okinawa Churashima Foundation
3. Activities and Findings
3.1 Metabolomic Analysis of Fission Yeast at the Onset of Nitrogen Starvation
To gain an understanding of the cellular response of the fission yeast S. pombe to nitrogen starvation at the metabolomic level, we performed a metabolome analysis of the cells over a short time-course immediately following nitrogen starvation. We focused on a time period of 1 h, before any obvious morphological changes in the cells can be observed. Changes during this time period would characterize the direct and immediate metabolic response to nitrogen starvation. To obtain metabolomic data sets, three independent wild-type S. pombe cultures were cultivated at 26°C in liquid EMM2 medium. Cells were collected by vacuum filtration and shifted to EMM2-N medium (EMM2 lacking NH4Cl) to induce nitrogen starvation. Samples were taken from each culture at the time points of 0 (corresponding to the vegetative culture before medium shift), 15, 30, and 60 min for metabolite detection using liquid chromatography on the ZIC-pHILIC column (Merck SeQuant, Umeå, Sweden) and the LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, USA). We identified 75 individual metabolite signals, mainly by comparing their m/z values and retention times with authentic pure standards.
The results showed that during this time-course, 6 metabolites (2-oxoglutarate, cytidine, hercynylcysteine sulfoxide, S-methyl-ergothioneine, succinate, and trehalose) increased more than 10-fold, and 11 metabolites (AICAR, aspartate, citruline, FGAM, FGAR, glutamine, histidine, IMP, PRPP, SAICAR, and thiamine) decreased more than 10-fold. Interestingly, the level of high-energy compounds, such as adenosine triphosphate (ATP), S-adenosyl methioneine (SAM), and nicotinamide adenine dinucleotide (NAD+) remained relatively stable during the time-course, suggesting that the cellular energy pool was not impaired during the initial stage of nitrogen starvation. Overall, peak areas of 29 of 75 identified metabolites (38.7%) remained within a two-fold change (Figure 1). The most significant changes occurred during the first 15 min, when trehalose, 2-oxoglutarate (2OG), and succinate increased, while purine biosynthesis intermediates (PRPP, FGAR, FGAM, SAICAR, AICAR and IMP) rapidly diminished. The following period (between 15 and 60 min after nitrogen starvation) was mainly characterized by changes in the amino acid composition. After passing the 15 min time point, all free amino acids sharply declined, although the levels of some modified amino acids, such as methylated lysine and arginine, gradually increased. Among these was also ergothioneine, a sulphur-containing derivative of trimethyl histidine. Besides ergothioneine itself, we also detected a sharp increase in an S-methyl derivative of ergothioneine, and hercynylcysteine sulfoxide, a known precursor of ergothioneine.
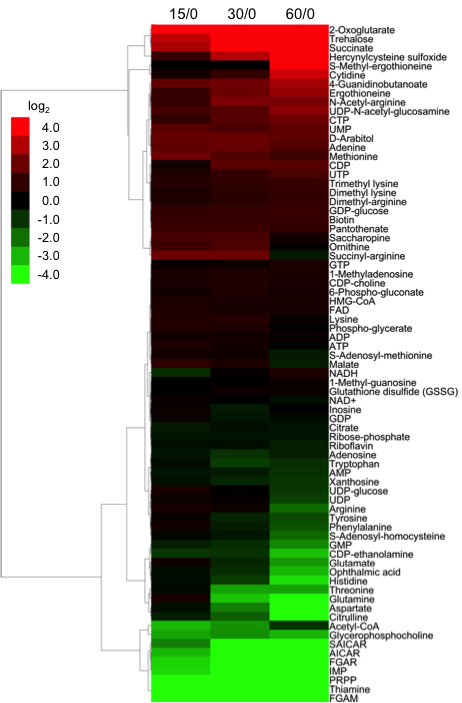
Figure 1. Time-course of changes in the peak areas of 75 metabolites.The time-course of changes in the peak areas of 75 metabolites in log2 values was clustered hierarchically by a complete linkage method using the Cluster 3.0 software ver. 1.50, and visualized as a tree view image by the Java TreeView software ver. 1.1.6r2.
In this study, we characterized the metabolic response of S. pombe to nitrogen starvation during the critical first hour, as schematized in Figure 2. The accumulation of trehalose and ergothioneine was also observed under glucose starvation in our previous study, so such response seems to be rather non-specific. On the other hand, the fast shut-off of purine biosynthesis and the sharp rise of 2OG and succinate seemed to be caused specifically by depletion of NH4Cl from the culture medium. 2OG is required for ammonia assimilation in S. pombe by glutamate dehydrogenase, and depletion of ammonia might thus lead to the accumulation of 2OG. The increase in succinate might be a direct consequence of 2OG accumulation, as 2OG can be converted to succinate by 2-oxoglutarate oxygenases. On the other hand, the decreases of intermediates in the purine biosynthesis pathway might implicate inhibition of the biosynthesis of nitrogen-rich compounds, like purines. The level change of such key metabolites may represent an important mechanistic step to trigger subsequent cellular regulations under nitrogen starvation.
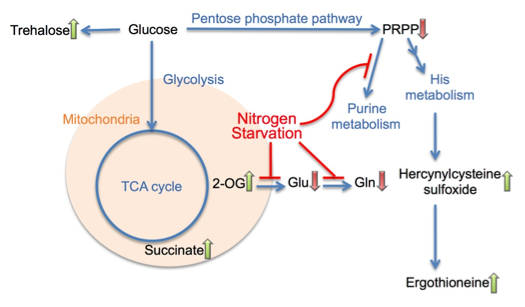
Figure 2. A summary diagram of metabolomic changes in S. pombe cells at the onset of nitrogen starvation.
3.2 Klf1, a C2H2 zinc finger-transcription factor, is required for cell wall maintenance during long-term quiescence in differentiated G0 phase
Previously, we reported several G0-specific-essential genes encoding the following proteins: two zinc-finger (designated zf) transcription factors Klf1 and Rsv2; two phosphoinositide-binding proteins involved in autophagy, Atg18 and Mug179/Atg18b; PP2A-like phosphatase inhibitor Sds23/Moc1 and PP2A-like phosphatase activator Ypa1/Rrd1; AMP-kinase regulatory subunit Cbs2; and GTPase-activating proteins Gyp7 and Spac589.07c (Shimanuki et al, 2007). In the present study, we investigated the role of Klf1, as its gene deletion mutant has a unique phenotype in two senses: the late timing of the appearance of defects in the G0 phase, and a drastically aberrant cell morphology. Klf1 is a Krüppel-like transcription factor with a 7-amino acid Cys2His2-type zinc finger motif. The deletion mutant, ∆klf1, normally divides in vegetative medium, but proliferation is not restored after long-term G0 quiescence. Cell biologic, transcriptomic, and metabolomic analyses revealed a unique phenotype of the ∆klf1 mutant in quiescence. Mutant cells had diminished transcripts related to signaling molecules for switching to differentiation; however, proliferative metabolites for cell-wall assembly and antioxidants significantly increased. Further, the size of ∆klf1 cells increased markedly during quiescence due to the aberrant accumulation of calcofluor-positive chitin-like materials beneath the cell wall (Figure 3). After 4 weeks of quiescence, reversible proliferation ability was lost, but metabolism was maintained. Klf1 thus plays a role in G0 phase longevity by enhancing the differentiation signal and suppressing metabolism for growth. If Klf1 is lost, S. pombe fails to maintain a constant cell size and normal cell morphology during quiescence.
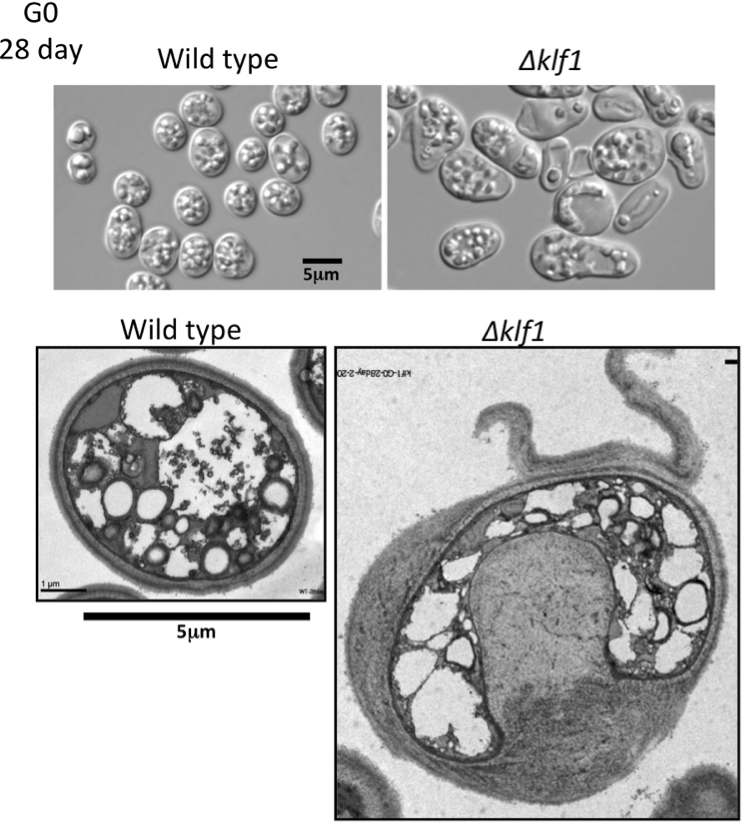
Figure 3. The size of ∆klf1 cells increased markedly during quiescence.
To date, among thousands of mutants searched by cytologic screening, we have found no other mutant with the same phenotype. The ∆klf1 mutants often had a 10–fold larger cell volume after a 4-week quiescent phase. Such an ‘obese’ cell phenotype might be considered a kind of senescence defect in non-dividing cells. Metabolomic data suggested that these quiescent mutant cells were still metabolically active, as the levels of nucleotide triphosphates were normal, although several individual metabolites greatly accumulated in the mutant (Figure 4). We thus concluded that the long-term G0 phenotype of the ∆klf1 mutant uniquely disrupts the homeostasis of quiescence, allowing for an increase in cell volume, one of the features of growing cells.
In summary, this study showed that Klf1 has a unique function to maintain cell size and cell morphology in G0 quiescence. Klf1 is implicated in chronologic cell longevity, homeostasis of non-dividing cells, cell wall maintenance, and oxidative stress response. Klf1 is thus an excellent model for understanding metabolic factors that govern cellular quiescence. This study was published in PLOS ONE (2013).
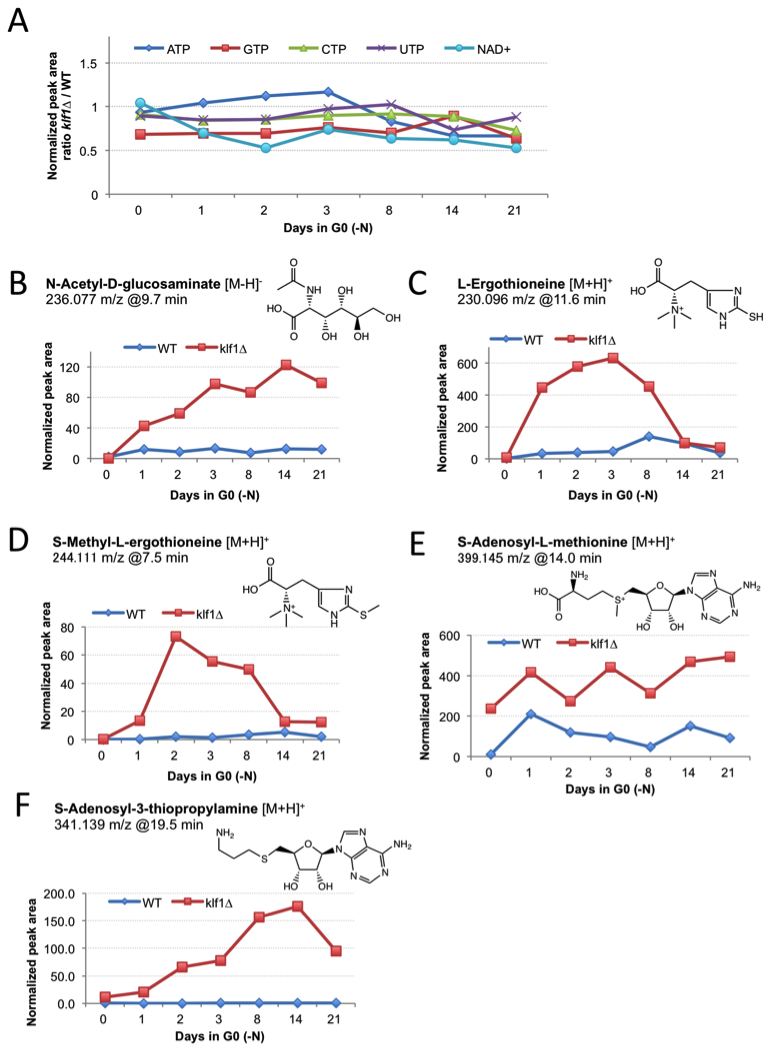
Figure 4. Comparison of metabolite profiles in wild type (WT) and ∆klf1 cells.
3.3 Study of ergothioneine biosynthetic genes in fission yeast
Ergothioneine is a small, sulfur-containing metabolite (229 Da) synthesized by various species of bacteria and fungi, which can accumulate to millimolar levels in tissues or cells (e.g., erythrocytes) of higher eukaryotes. Based on the reported accumulation of ergothioneine under glucose starvation (Pluskal et al, 2011) and nitrogen starvation (Sajiki et al, 2013), as well as in the ∆klf1 mutant (Shimanuki et al, 2013), we extensively investigated the biosynthesis of this compound in fission yeast. We identified the first biosynthetic gene, egt1+, by sequence homology with previously published genes from Neurospora crassa and Mycobacterium smegmatis. The second step of the biosynthesis, represented by the egt2+ gene, was identified by screening several homologs of the corresponding mycobacterial enzyme. The complete pathway is summarized in Figure 5. We expect the identification of these genes will allow us to study the function of this compound under cellular quiescence.
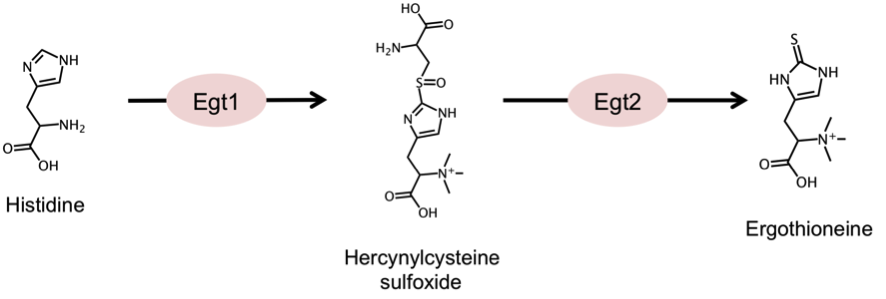
Figure 5. Summary of the ergothioneine biosynthetic pathway in S. pombe
3.4 Identification of Mis18-interacting factors
CENP-A is a centromere-specific variant of histone H3 that is required for accurate chromosome segregation. Centromere localization of CENP-A requires Mis16 and Mis18, which together form a complex. The Mis18 complex is hypothesized to have a role in centromere priming for CENP-A deposition in the subsequent cell cycle, and thus histone acetylation is suggested to be involved in Mis18 complex function (Fujita et al. 2007). The molecular function of the Mis18 complex, however, remains unclear. We investigated the molecular organization of the Mis16-Mis18 complex in fission yeast and identified two novel Mis18-interacting proteins, Mis19 and Mis20 (Figure 6). In addition, we screened extragenic suppressors of mis18 and mis19 mutants, and identified the functional link between the Mis18 complex and nonsense-mediated mRNA decay factors or SWI/SNF chromatin- remodeling complex.
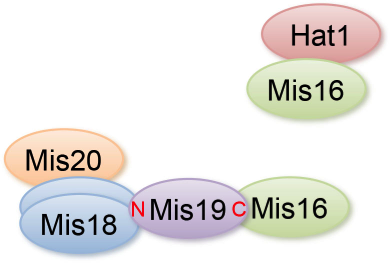
Figure 6. Summary of the physical interactions of Mis16- and Mis18-interacting proteins.
3.5 R&D cluster projects in Okinawa
During a 3-year period (FY2011-2013), we participated in the Okinawa Prefecture Intellectual Cluster Research Project, for future commercial development of health and medical care in Okinawa. Two laboratories at the Ryukyu and Kyoto Universities, and two private companies, SOMNOQUEST and Sentan Iryou Kaihatsu, joined together to form a research team that will use an analytic technology called “metabolome analysis” to search for Okinawan products that contain substances to treat disease, and to conduct research on the transdermal absorption process of such substances.
In collaboration with Kyoto and Ryukyu universities, the R&D cluster project ‘Metabolomic study for the technologic development of health-longevity improvement through new approaches to the transdermal absorption of active ingredients’ in Okinawa was continued in FY2013. Our developed protocol for obtaining blood metabolome samples (Figure 7) as well the method for data analysis were published in a Japanese review article, Rinsho Ketsueki (Chaleckis et al 2013). In human blood, we can detect more than 100 compounds verified by purchased standards. In addition to known compounds, we found several novel blood metabolites. Comparison of young (22-37 years, 15 persons) and old (70-96 years, 15 persons) blood metabolomes revealed several identified compounds as well as unidentified peaks, revealing differences in levels and modes of distribution between the age groups. Many of the compounds detected in human blood are found also in fission yeast, enabling us to dissect their role using powerful yeast genetic methods.
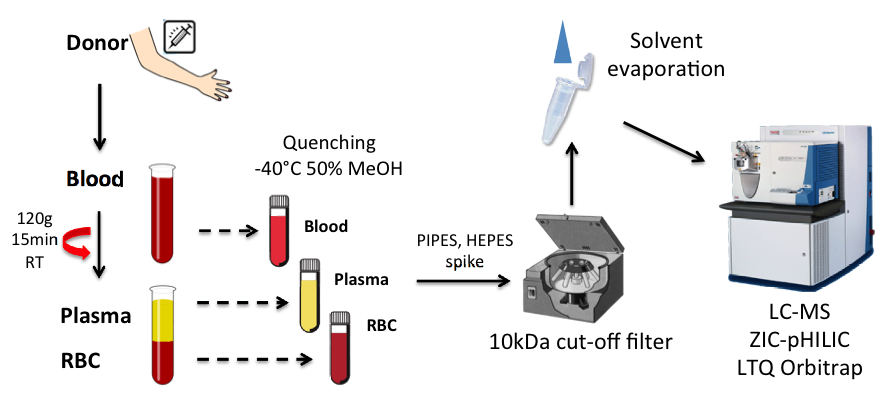
Figure 7. Schema of human blood metabolome sample preparation. Metabolic compounds were extracted in 50% MeOH at -40°C from whole blood, plasma, and RBC separated by centrifugation. Extracted metabolites were isolated using a 10k-Da cut-off filter, concentrated, and analyzed on a LC-MS system.
In collaboration with SOMNOQUEST and Kyoto University, we examined the metabolism of active ingredients and their effect on mouse blood metabolism after oral or percutaneous administration of Okinawan medicinal plants or pure anti-oxidants. We investigated the possibility of delivering active compounds though skin using a mouse model. We successfully achieved oral and skin delivery of an anti-oxidant, resveratrol, in hairless mice. Resveratrol and its metabolites were detected in different tissues; metabolism models were proposed after oral and skin administration (Figure 8). Yomogi (Japanese mugwort) extracts were applied on mouse skin or delivered orally. We detected several extract components as well as their metabolites in various mouse tissues after administration. Skin absorption is a feasible alternative method of delivering compounds into the body.
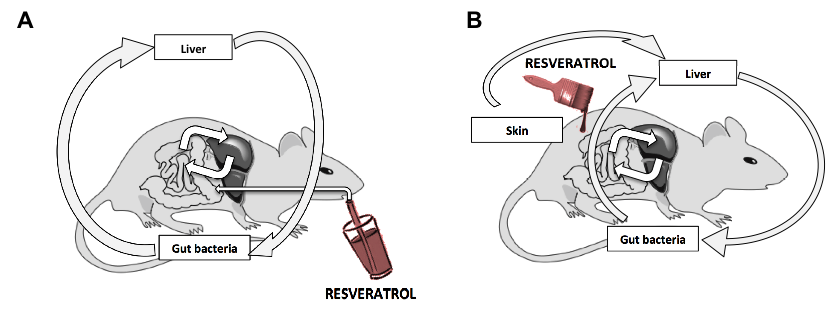
Figure 8. Resveratrol metabolism after oral administration (A) or skin absorption (B)
4. Publications
4.1 Journals
- Earnshaw, W. C., Allshire, R. C., Black, B. E., Bloom, K., Brinkley, B. R., Brown, W., Cheeseman, I. M., Choo, K. H., Copenhaver, G. P., Deluca, J. G., Desai, A., Diekmann, S., Erhardt, S., Fitzgerald-Hayes, M., Foltz, D., Fukagawa, T., Gassmann, R., Gerlich, D. W., Glover, D. M., Gorbsky, G. J., Harrison, S. C., Heun, P., Hirota, T., Jansen, L. E., Karpen, G., Kops, G. J., Lampson, M. A., Lens, S. M., Losada, A., Luger, K., Maiato, H., Maddox, P. S., Margolis, R. L., Masumoto, H., McAinsh, A. D., Mellone, B. G., Meraldi, P., Musacchio, A., Oegema, K., O'Neill, R. J., Salmon, E. D., Scott, K. C., Straight, A. F., Stukenberg, P. T., Sullivan, B. A., Sullivan, K. F., Sunkel, C. E., Swedlow, J. R., Walczak, C. E., Warburton, P. E., Westermann, S., Willard, H. F., Wordeman, L., Yanagida, M., Yen, T. J., Yoda, K. & Cleveland, D. W. Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 21, 101-106, doi:10.1007/s10577-013-9347-y (2013).
- Sajiki, K., Pluskal, T., Shimanuki, M. & Yanagida, M. Metabolomic Analysis of Fission Yeast at the Onset of Nitrogen Starvation. Metabolites 3(4), 1118-1129 (2013).
- Shimanuki, M., Uehara, L., Pluskal, T., Yoshida, T., Kokubu, A., Kawasaki, Y. & Yanagida, M. Klf1, a C2H2 zinc finger-transcription factor, is required for cell wall maintenance during long-term quiescence in differentiated G0 phase. PLoS One 8, e78545, doi:10.1371/journal.pone.0078545 (2013).
- Chaleckis, R., Pluskal, T., Hayashi, T., Kondoh, H. & Yanagida, M. in Rinsho Ketsueki Vol. 54 臨床血液 57-68 (2013).
4.2 Books and Other One-Time Publications
Nothing to report
4.3 Oral and Poster Presentations
- Yanagida, M. Control of quiescence induced by starvation of glucose and nitrogen source in Joint Conference of HGM2013 and 21st International Congress of Genetics' Genetics & Genomics of global health and sustainability, Marina Bay Sands, Singapore (2013).
- Yanagida, M. Approaches to health, disease and aging by comprehensive blood metabolomics, in Speacial forum; Health, longevity, life innovation, Nakanoshima, Osaka (2014).
- Yanagida, M. Life Science in Japan- Past 50 years and its future, in The 91st Annual Meeting of the Physiological Society of Japan, Kagoshima Univ., Kagoshima (2014).
- Nakazawa, N., Sajiki, K., Xu, X. & Yanagida, M. ‘Transcriptsbuster’ in mitosis?: Condensin resolves the obstructive effect of mitotic transcripts on chromosome segregation, Cold spring harbor laboratory meeting 'Cell Biology of Yeast' (2013).
- Pluskal, T. Ergothioneine biosynthesis pathway in S. pombe revealed by metabolomic analysis, in 7th International Fission Yeast Meeting, POMBE2013, London, UK (2013).
- Yanagida, M. How do fission yeast cells respond to low glucose concentrations and proliferate?, in 7th International Fission Yeast Meeting, POMBE2013, London, UK (2013).
- Nakazawa, N. Fission Yeast condensin ensures sister chromatid separation by relieving the obstructive effect of transcription on segregation, in The 46th Yeast Genetic Forum Sendai, Japan (2013).
- Sajiki, K. Rescue screening of a temperature sensitive mutant library by rapamycin, in The 3rd TOR meeting, Okazaki, Japan (2013).
- Yanagida, M. Metabolome analysis of human blood, plasma and RBC (red blood cell): What we can learn from individual differences in metabolomic data?, in The 75th Annual Meeting of the Japanese Society of Hematology, Sapporo, Hokkaido (2013).
- Yanagida, M. Model study of diabetes mellitus: How cells respond to the change in glucose concentration in the culture medium?, in Tohoku Medical Society Symposium, Tohoku University, Sendai (2013).
- Yanagida, M. Metabolomic study for the technological development of health-longevity improvement through new approaches to the transdermal absorption of the active ingredients, in Okinawa Intellectual Cluster Symposium, Naha, Okinawa (2013).
- Hayashi, T., Ebe, M., Nagao, K., Kokubu, A. & Yanagida, M. Identification of novel centromere proteins Mis19 and Mis 20 as Mis18-interacting partners in S. pombe, in 7th International Fission Yeast Meeting London, UK (2013).
- Hayashi, T., Pluskal, T. & Yanagida, M. Impaired phosphatidylcholine synthesis and aberrant nuclear division in mutants of SAM and SAH cycle enzymes, in 7th International Fission Yeast Meeting, POMBE2013, London, UK (2013).
- Mehrotra, R., Mori, A., Saitou, S. & Yanagida, M. Screening of genes that regulates the localization of Ght5 in low glucose sensitive mutants, in Okinawa Intellectual Cluster Symposium, Naha, Okinawa (2013).
- Nakazawa, N., Villar-Briones, A., Sajiki, K., Arakawa, O., Xu Xingya & Yanagida, M. Condensin de novo Accumulation at the Transcriptional Active Genes from Metaphase to Anaphase, in 7th International Fission Yeast Meeting, POMBE2013, Lonodon, UK (2013).
- Pluskal, T. Highly accurate chemical formula prediction tool utilizing high-resolution mass spectra, MS/MS fragmentation, heuristic rules, and isotope pattern matching, in 9th International Conference of the Metabolomics Society, Glasgow, Scotland (2013).
- Pluskal, T. Ergothioneine biosynthesis pathway in Schizosaccharomyces pombe revealed by metabolomic analysis, in Okinawa Intellectual Cluster Symposium, Naha, Okinawa (2013).
- Yanagida, M., Akai, Y. & Nakazawa, N. Autophosphorylation of condensin SMC’s subunits Cut3 and Cut14; Revelation of a part of the ATP cycle, in 7th International Fission Yeast Meeting, POMBE2013, London, UK (2013).
- Yanagida, M., Shimanuki, M. & Pluskal, T. Klfl, a C2H2 zinc finger-transcription factor, is required for cell volume maintenance in the fission yeast G0 phase, in 7th International Fission Yeast Meeting, POMBE2013, London, UK (2013).
- Ebe, M., Teruya, T., Chaleckis, R., Pluskal, T., Kondoh, H. & Yanagida, M. Presentation Goat and dolphin blood Metabolome analysis, in Okinawa Intellectual Cluster Symposium, Naha, Okinawa (2013).
- Hayashi, T. Aberrant nuclear division in mutants of methionine cycle enzymes, in The 46th Yeast Genetic Forum Sendai, Japan (2013).
- He, X., Ebe, M., Pluskal, T., Chaleckis, R. & Yanagida, M. To find the hallmarks of anti-aging by mouse red blood cell proteome in Okinawa Intellectual Cluster Symposium, Naha, Okinawa (2013).
- Sajiki, K. Metabolomic analysis of fission yeast at the onset of nitrogen starvation, in Okinawa Intellectual Cluster Symposium, Naha, Okinawa (2013).
- Xu, X. Comprehensive isolation of ts and/or drug sensitive mutants for condensin’s non-SMC subunits Cnd1 and Cnd1 using error-prone PCR, in The 46the Yeast Genetic Forum, Sendai, Japan (2013).
- Nakazawa, N. Condensin ensures sister chromatid separation at transcriptionally-activated loci by relieving the obstructive effect of transcription on chromosome segregation, Oxford University, Dunn School of Pathology, UK (2013).
- Yanagida, M. Control of chromosome segregation and cell quiescence longevity, The 29th Steinberg/ Wylie Lecture in Biochemistry, The University of Maryland School of Medicine, USA (2013).
- Yanagida, M. All-out changes in cell regulations after low glucose shift, Oxford University, UK (2013).
- Yanagida, M. Genetic control of glucose starvation response in fission yeast, Biozentrum, Basel (2013).
- Yanagida, M. Clearing Mitosis and the Role of Condensin, Zurich Institut fur Biochemie, Swiss (2013).
- Yanagida, M. Future Prospects of Human Blood Metabolome Analysis in Health and Medical Science, OIST, Okinawa (2013).
- Yanagida, M. Approaches by metabolomic analyses for starvation response of yeast and toward understanding human longevity, Shimane University (2013).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
Nothing to report



