FY2011 Annual Report
G0 Cell Unit
Professor Mitsuhiro Yanagida

Abstract
Regulation of cellular proliferation and quiescence (G0 phase) in response to changes in the environment, such as the availability of nutrients, is a central issue in biology and medicine. Our research goal is to elucidate the molecular mechanisms that regulate pivotal events in proliferation and quiescence, such as chromosome segregation, coupling of cell growth and division, and cellular adaptation to nutrient paucity. For our studies, we use the fission yeast Schizosaccharomyces pombe, a genetically-tractable simple unicellular eukaryote, as a model organism. In FY2011, we further evaluated the G0 phase induced by nitrogen starvation and the adaptation of S. pombe to glucose-limited conditions. In addition, we investigated various regulatory mechanisms of proliferation. We published 1 review article and 10 original research articles on metabolomic studies under glucose starvation, regulation of growth and mitosis by Target of Rapamycin (TOR) complexes, chromosome condensation in mitosis, regulation of the proteasome, and other topics, as introduced in 3. Activities and Findings.
1. Staff
- Dr. Takeshi Hayashi, Group leader
- Dr. Kojiro Takeda, Group leader
- Dr. Takahiro Nakamura, Researcher (until November)
- Dr. Kenichi Sajiki, Researcher
- Dr. Norihiko Nakazawa, Researcher
- Ms. Risa Uehara, Technical staff
- Ms. Ayaka Mori, Technical staff
- Mr. Tomas Pluskal, Technical staff
- Mr. Alejandro Villar-Briones, Technical staff
- Mr. Masahiro Ebe, Technical staff
- Ms. Yuria Tahara, Technical staff
- Ms. Yuko Akai, Technical staff
- Ms. Tomomi Teruya, Research administrator (until September)
- Ms. Chikako Sugiyama, Research administrator (from September)
- Ms. Eulaia Lopez-Galcia, Research assistant
2. Collaborations
- Theme: Metabolite structure determination using NMR
- Type of collaboration: Joint research
- Researchers:
- Professor Masar Ueno, Department of Molecular Biotechnology, Graduate school of Advanced Science of Matter, Hiroshima University
- Theme: Identification of fission yeast mutants, which show growth defect under low glucose condition
- Type of collaboration: Joint research
- Researchers:
- Professor Shigeaki Saitoh, Institute of Life Science, Kurume University
- Theme: Identification of the molecular mechanism, required for the maintenance of cell viabiity in low glucose condition
- Type of collaboration: Joint research
- Researchers:
- Professor Kunihiro Ohta, Department of Life Science, Graduate School of Arts and Science, University of Tokyo
- Theme: Metabolomic analysis of human blood
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Takumi Mikawa, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Research for DNA metabolism in fission yeast
- Type of collaboration: Joint research
- Researchers:
- Professor Tomohiro Matsumoto, Radiation Biology Center, Kyoto University
3. Activities and Findings
3.1 Comprehensive screening of genes required for maintenance of the G0/quiescent phase
Transition from the growth phase to the G0 phase is associated with extensive changes in cellular behavior and structure. The genes essential for establishing and maintaining G0 phase, however, are largely unknown. The fission yeast S. pombe is an excellent model for studying this problem because these cells enter the G0 phase under nitrogen starvation. We screened our temperature-sensitive mutant library for strains with defective survival in the G0 phase induced by nitrogen starvation, and identified 183 strains among the 818 strains tested. From these 183 strains, we identified 86 unique genes responsible for G0 phase survival by whole-genome sequencing. In recent years, deletion mutant library sets have also become commercially available. We used the Bioneer haploid deletion sets Ver. 1 to 3 to identify genes essential for the G0 phase. By spot testing and viability screening under nitrogen starvation, we analyzed more than 3700 deletion strains. We utilized two laboratory automation systems to handle the number of strains. These screenings led us to identify 102 strains with considerably decreased viability within 4 weeks after nitrogen starvation. The deleted genes of these strains covered various cellular functions, such as vesicle trafficking including autophagy, stress response signaling, chromatin remodeling, and RNA processing. We also observed the morphology of these strains in G0 phase under a light microscope, and observed an interesting morphology for each group of strains having mutations in genes with similar functions.
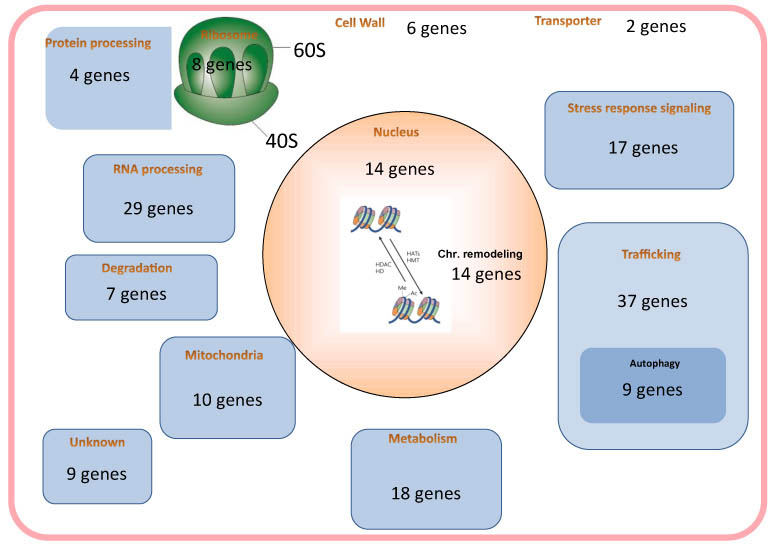
Figure 1. Essentials genes for the maintenance of G0 phase
3.2 S. pombe metabolomic analysis under low glucose conditions
In April 2011, our study on the cell division-quiescence behavior of the fission yeast S. pombe under limited glucose conditions was published in FEBS Journal. We described the detailed phenotypes of cells fixed in a microfluidic perfusion system and supplied with a constant amount of glucose in the culture medium. Comprehensive metabolomics analysis was performed using liquid chromatography-mass spectrometry (LC-MS) and marker metabolites were established for characteristic phenotypes (Figure 2). Based on the cellular and metabolic phenotypes characterized, we designated the different glucose concentrations as excess (111 mM), regular (11.1 mM), diet (4.4 mM), severe diet (2.2 mM), sub-starvation (1.7 mM), starvation (1.1 mM), and fasting (0 mM), as summarized in Table 1. Furthermore, we analyzed the genome stability under low glucose conditions and provided evidence that cellular adaptation to glucose starvation is linked to resistance to oxidative stress.

Figure 2. A to D. Peak areas of potential biomarker compounds in 10 different glucose concentrations determined by the LC/MS method. E to G. Time course (8 h) analysis of several critical metabolites upon entering the fasting condition (0 mM glucose in the medium). H to J. Time course (7 d) analysis of the peak areas of characteristic compounds upon entering the starvation condition (1.1 mM glucose, 100x diluted from the standard EMM2 medium).
|
Glucose content |
Designated terminology |
Biomarker metabolites |
Cell division |
||
|
mM |
w/v (%) |
mg/dl |
|||
|
111 |
2 |
2,000 |
Excess* |
Biotin ↑ |
Regular |
|
11.1 |
0.2 |
200 |
Regular |
Biotin ↑ |
Regular |
|
4.4 |
0.08 |
80 |
Diet |
Trehalose ↑, ergothioneine ↑ |
Regular, cell length is reduced |
|
2.2 |
0.04 |
40 |
Severe diet |
Fructose-1,6-diphosphate ↓ |
Stochastic |
|
1.7 |
0.03 |
30 |
Sub-starvation |
|
Stochastic |
|
1.1 |
0.02 |
20 |
Starvation |
S-adenosyl-methionine ↑ |
Only occasional division |
|
0 |
0 |
0 |
Fasting |
CDP-choline ↑, ATP ↓, S-adenosyl-homocysteine ↑ |
No division |
Table 1. Summary of the glucose concentrations analyzed in this work. Concentrations are listed in three commonly used notations: millimolar (mM), percentage (w/v), or mg/dl. Biomarker metabolites increased (↑) or decreased (↓) in each condition are noted, as well as the corresponding cell division phenotypes.
3.3 Opposite, but coordinated, roles of Tor2 and Tor1 kinases in growth and mitosis
TORCs (Target of Rapamycin Complexes), which are vital for nutrient utilization, contain a catalytic subunit with the phosphatidyl inositol kinase-related kinase (PIKK) motif. TORC1 is required for cell growth, while the functions of TORC2 are less well understood. To better understand the distinct functions of TORC1 and TORC2, we used the previously isolated TORC1 mutant tor2-L2048S in the phosphatidyl inositol kinase (PIK) domain and a new TORC2 mutant tor1-L2045D, which harbors a mutation at the same site. These fission yeast mutants allowed us to critically compare TORC2 functions with TORC1 functions in growth and in the cell cycle. While mutated TORC1 and TORC2 displayed diminished kinase activity and FKBP12/Fkh1-dependent rapamycin sensitivity, their phenotypes were nearly opposite in mitosis. Premature mitosis and G2-M delay occurred in TORC1 and TORC2 mutants, respectively (Figure 3A). The fission yeast TORC2 has a cell cycle role through determining the proper timing of Cdc2 Tyr15 dephosphorylation and cell size under limited glucose conditions, whereas TORC1 restrains mitosis. Separase/Cut1 is a conserved protease that is essential for anaphase progression. Securin/Cut2 is a chaperone-inhibitor for Cut1 and is degraded in the transition from metaphase to anaphase by the anaphase-promoting complex (APC)/cyclosome. Surprisingly, separase/cut1-securin/cut2 mutants were rescued by the TORC1/tor2-L2048S mutation, rapamycin addition, or even Fkh1 deletion, whereas these mutants showed synthetic defects with TORC2/tor1-L2045D (Figure 3B). TORC1 and TORC2 coordinate growth, mitosis, and cell size control, as Wee1 and Cdc25 do for the entry into mitosis. This study was published in Open Biology (Ikai et al., 2011).

Figure 3. Phenotypic differences between tor1-D and tor2-S. A. The cell length (micrometers, columns) and the cell number (line) of wild-type (black), tor1-D (red) and tor2-S (yellow) were measured for cells cultured initially at 26˚C and then shifted to 30˚C, 33˚C and 37˚C for 0-8 h. DAPI-stained cells taken at 8 h are also shown. B. Synthetic rescue of cut1 or cut2 mutant by tor2-S and additive ts phenotype of the double mutants between tor1-D and cut1-693 or cut2-364.
3.4 Opposing roles of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing
Condensin is required for chromosome dynamics and diverse DNA metabolism. Condensin contains two SMC (Structural Maintenance of Chromosomes) subunits with the terminal globular domains connected to a coiled-coil that is interrupted by a central hinge. We identified a novel fission yeast SMC-hinge mutant, cut14-Y1, which shows defects in DNA damage repair and chromosome segregation (Akai et al., 2011). The cut14-Y1 mutant contains an amino acid substitution at a conserved hinge residue of Cut14/SMC2, resulting in diminished DNA binding and annealing. A replication protein A mutant, ssb1-418, greatly alleviated the repair and mitotic defects of cut14-Y1. Ssb1 protein bound to single-strand DNA was removed by condensin or the SMC dimer through DNA reannealing in vitro. Thus, condensin may wind up DNA strands to unload chromosomal components after DNA repair and prior to mitosis (Figure 4).
We also critically examined whether condensin continuously acts on chromosomes during mitosis and compared its role with that of DNA topoisomerase II (Top2). The results established that mitotic chromosomes require condensin and Top2 throughout mitosis, and even in telophase (Nakazawa et al., 2011). We also demonstrated that Cnd2/Barren is the target subunit of aurora-B like kinase Ark1. Condensin may thus have to keep apart from the segregated chromosomes in telophase.
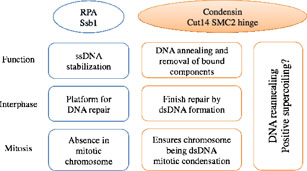
Figure 4. A diagram depicting the relationship between the condensin SMC Cut14 hinge and Ssb1
3.5 Studies of the factors affecting proteasomal function and localization of the proteasome
Ubiquitin/proteasome-dependent proteolysis has pivotal roles in many essential cellular processes, such as cell cycle control, transcriptional regulation, and protein quality control. We previously studied the roles of the proteasome in G0 phase maintenance and reported that maintenance of mitochondria by the proteasome and autophagy may be important for cell survival in the G0 phase. To further our understanding of proteasome function, we performed a genome-wide screening of S. pombe mutants showing hypersensitivity to Bortezomib/Velcade/PS-341, a chemical proteasome inhibitor that is used for anti-cancer therapy. Among the 2815 genes screened, we identified 19 genes whose deletions induce strong synthetic lethality with Bortezomib. We designated these genes SLB (Synthetic Lethal with Bortezomib) genes (Table 2). Thirteen identified genes are conserved in humans and are thus potentially interesting for further studies in human culture cells. If the same synthetic lethality effects occur in human cells, such SLB genes might be useful for the development of new therapies or diagnostic methods. These findings were published in 2011.
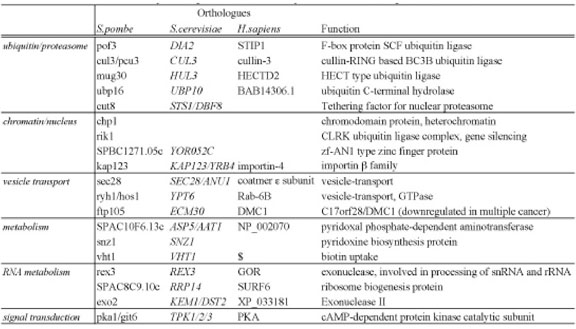
Table 2. The list of SLB (Synthetic Lethal with Bortezomib) genes
In S. pombe, the proteasome localizes to the nucleus, especially the nuclear envelope and chromatin region, depending on the function of Cut8 protein. The molecular mechanism of Cut8, however, is not fully understood. To obtain mechanistic insights into Cut8 function, the crystal structure of Cut8 was solved in collaboration with Dr. Maria Schumacher at Duke University (Figure 5). Cut8 comprises three domains, the N-terminal proteasome binding domain, homo-dimerization domain, and 14-3-3-like domain. The 14-3-3 protein binds phosphorylated protein and negatively charged membranes. The 14-3-3-like domain of Cut8 is essential for Cut8 localization to the nuclear envelope, Cut8-lipid membrane association in vitro, and proteasome enrichment in the nucleus, suggesting that an interaction between Cut8 and the proteasome near the nuclear envelope, which depends on the 14-3-3 like domain of Cut8, is important for proper nuclear localization of the proteasome.

Figure 5. The structure of Cut8, tethering factor for the nuclear proteasome
3.6 Other activities
Comprehensive screening of mutants showing growth defects under glucose-limited conditions was performed and functional analyses on isolated mutants are in progress (in collaboration with Kurume University).
In FY2011, our research proposal, ‘Metabolomic study for the technological development of health-longevity improvement through new approaches to the transdermal absorption of the active ingredients’, was launched as one of the R&D cluster projects in Okinawa. Two laboratories at the Ryukyu and Kyoto Universities, and two private companies, SOMNOQUEST and Sentan Iryou Kaihatu, joined to form a research team that will use an analytic technology called “metabolome” to search for Okinawan products that contain substances to treat disease, and to conduct research on the transdermal absorption process of such substances. The team will also conduct genetic analyses to investigate why the people of Okinawa are the longest living people in the world..
4. Publications
4.1 Journals
- Ikai, N., Nakazawa, N., Hayashi, T., Yanagida, M. The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in Schizosaccharomyces pombe. Open Biology 1, 110007--110007 doi:10.1098/rsob.110007 (2011).
- Kojiro Takeda, Ayaka Mori, and Mitsuhiro Yanagida. Identification of Genes Affecting the Toxicity of Anti-Cancer Drug Bortezomib by Genome-Wide Screening in S.pombe. PLoS One 6, e22021 (2011).
- T. Pluskal , T. Hayashi , S. Saitoh , A. Fujisawa and M. Yanagida., (2011) Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. FEBS J 278 1299--1315
- Takahashi, Katsunori , Imano, Ryota , Kibe, Tatsuya , Seimiya, Hiroyuki , Muramatsu, Yukiko , Kawabata, Naoki , Tanaka, Genki , Matsumoto, Yoshitake , Hiromoto, Taisuke , Koizumi, Yuka , Nakazawa, Norihiko , Yanagida, Mitsuhiro , Yukawa, Masashi , Tsuchiya, Eiko and Ueno, Masaru, (2011) Fission yeast Pot1 and RecQ helicase are required for efficient chromosome segregation. Molecular and cellular biology (31)3 495-506 doi: 10.1128/MCB.00613-10
- Kiyomitsu, T. , Murakami, H. and Yanagida, M., (2011) Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Molecular and Cellular Biology 5 998-1011 doi: 10.1128/MCB.00815-10
- Y. Shiroiwa , T. Hayashi , Y. Fujita , A. Villar-Briones , N. Ikai , K. Takeda , M. Ebe and M. Yanagida., (2011) Mis17 Is a Regulatory Module of the Mis6-Mal2-Sim4 Centromere Complex That Is Required for the Recruitment of CenH3/CENP-A in Fission Yeast. PLoS One 6 e17761
- N. Nakazawa, R. Mehrotra, M. Ebe and M. Yanagida., (2011) Condensin phosphorylated by the Aurora-B-like kinase Ark1 is continuously required until telophase in a mode distinct from Top2. J Cell Sci 124 1795--1807
- M. Shimura, Y. Toyoda, K. Iijima, M. Kinomoto, K. Tokunaga, K. Yoda, M. Yanagida, T. Sata and Y. Ishizaka (2011) Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation. J Cell Biol doi: 10.1083/jcb.201010118
- K. Takeda, Tonthat, N. K., Glover, T., Xu, W., Koonin, E. V. , Yanagida, M. and Schumacher, M. A., (2011) Implications for proteasome nuclear localization revealed by the structure of the nuclear proteasome tether protein Cut8. Proc Natl Acad Sci U S A 108(41) 16950--16955 doi: 10.1073/pnas.1103617108
- Yanagida, M., Ikai, N., Shimanuki, M. and Sajiki, K., (2011) Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Philos Trans R Soc Lond B Biol Sci 366(1584) 3508--3520 doi: 10.1098/rstb.2011.0124
- Akai, Y., Kurokawa, Y., Nakazawa, N., Tonami-Murakami, Y., Suzuki, Y., Yoshimura, S. H., Iwasaki, H., Shiroiwa, Y., Nakamura, T., Shibata, E. and Yanagida, M. (2011) Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biology 1(4) 110023--110023 doi: 10.1098/rsob.110023
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- Kojiro Takeda, Nam K, Tonthat, Tiffany Glover, Weijun Xu, Eugene V. Koonin, Mitsuhiro Yanagida and Maria Schumacher. The nuclear proteasome tethering factor Cut8 directly binds to the nuclear envelope through 14-3-3-like structure, The 34th Annual Meeting of the Molecular Biology Society of Japan, Yokohama, Japan, Dec 13-16, 2011
- Pluskal, T., Hayashi, T., Saitoh, S., Fujisawa, A., Yanagida, M. Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast, 6th Japan-UK Cell Cycle Workshop, Windermere, United Kingdom, Apr 10-14, 2011
- Romanas Chaleckis, Tomáš Pluskal, Ebe Masahiro, Hiroshi Kondoh and Mitsuhiro Yanagida. Metabolomic comparison of human blood and fission yeastas a tool of longevity research, Symposium on Establishing a research hub toward the development of an intellectual cluster), Okinawa Industry Support Center, Naha, Okinawa, Dec 20, 2011
- Sajiki, K. Protein phosphatase regulators are essential for cellular quiescence in S.pombe, EMBO conference series: Europhosphatases 2011, Vienna , Austria, July 18-23, 2011
- Shigeaki Saitoh, Ayaka Mori, Nobuyasu Ikai, Takahiro Nakamura, Fumie Masuda, Saeko Soejima, Lisa Uehara, Mitsuhiro Yanagida. Systematic identification of fission yeast lgs genes required for cell proliferation under glucoselimited conditions, The 34th Annual Meeting of the Molecular Biology Society of Japan, Yokohama, Japan, Dec 13, 2011
- T. Pluskal, T. H., S. Saitoh, A. Fujisawa, M. Yanagida. Metabolomic analysis of stochastic division and quiescence under limited glucose, The 6th International Fission Yeast Meeting, Boston, June 28, 2011
- Yanagida, M. Control of mitosis by metabolites, nutrient restraint and DNA clearing, 6th Japan-UK Cell Cycle Workshop, Windermere, United Kingdom, Apr 10-14, 2011
- Yanagida, M. Cell size control: The meeting point of growth and cell division cycle, The 6th International Fission Yeast Meeting, Boston, June 25, 2011
- Yanagida, M. Protein dephosphorylation that restrains cell division under limited glucose, EMBO conference series: Europhosphatases 2011, Vienna , Austria, Jul 18-23, 2011
- YANAGIDA, M. Clearaing Mitosis and the Role of Condensin, Medical School, Peking University, Beijing, Dec 1, 2011
- YANAGIDA, M. New Sciences for aging and longevity, Main Campus, Peking University, Beijing, Dec 3, 2011
- YANAGIDA, M. Approaches for Human Longevity and Aging using Fission Yeast, Molecular Biology Of Yeast International Conference <YEAST2011>, Indian Institute of Technology Bombay (IITB), INDIA, Dec 10, 2011
- YANAGIDA, M. Translation: Technological Development for Metabolomic Study of Human Blood, Symposium on Establishing a research hub toward the development of an intellectual cluster, Okinawa Industry Support Center, Naha, OKINAWA, Dec 20, 2011
- YANAGIDA, M. Outline of joint research project, Symposium on Establishing a research hub toward the development of an intellectual cluster, Okinawa Industry Support Center, Naha, OKINAWA, Dec 20, 2011
- Yanagida, M. Approaches for Human Longevity and Aging using Fission Yeast, Seminar at Tokyo Metropolitan Institute of Medical Science, Tokyo, 02.15, 2012
- Yanagida, M. Clearing Mitosis and the Role of Condensin, Seminar at Riken, Saitama, Feb 16, 2012
- Yanagida, M. Metabolome analysis for the transdermal absorption of the active ingredients, the 2nd Committee on Establishing a research hub toward the development of an intellectual cluster, Okinawa, Feb 29, 2012
- Norihiko Nakazawa, Nobuyasu Ikai, Alejandro Villar-Briones, Takeshi Hayashi, and Mitsuhiro Yanagida. Are the chromosome segregation regulatory factors controlled by nutritional environment?), The 29th Chromosome Workshop, Hotel New Mitoya, Sendai, Jan 25-27, 2012
- Takeshi Hayashi, K. N., Aya Kokubu, Masahiro Ebe, Yohta Fujita and Mitsuhiro Yanagida. Identification of novel components of Mis18 complex required for CENP-A localization, The 29th Chromosome Workshop, Hotel New Mitoya, Sendai, Jan 25-27, 2012
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 The 5th International Workshop on Cell Regulations in Division and Arrest
- Date: Oct. 23~27, 2011
- Venue: OIST Campus
- Speakers:
- Jürg Bähler (University College London, UK)
- Simon Chan (University of California-Davis, USA)
- Claudio De Virgilio (University of Fribourg, Switherland)
- Meng-Qiu Dong (National Institute of Biological Science, China)
- Li-Lin Du (National Institute of Biological Science, China)
- Eric Greene (Columbia University, USA)
- Yoshie Harada (Kyoto University)
- Atsushi Hirao (Kanazawa University)
- Shigeaki Kato (University of Tokyo)
- Stephen Kearsey (University of Oxford, UK)
- Takehiko Kobayashi (National Institute of Genetics)
- Hiroshi Kondoh (Kyoto University)
- Valter Longo (University of Southern California, USA)
- Peter McHugh (University of Oxford, UK)
- Toru Miyazaki (University of Tokyo)
- Yoichi Nabeshima (Kyoto University)
- Keiichi Nakayama (Kyushu University)
- Keiko Nakayama (Tohoku University)
- Jonathon Pines (University of Cambridge, UK)
- Randy Poon (Hong Kong University of Science and Technology)
- Paul Russell (Scripps Research Institute, USA)
- Mitsutoshi Setou (Hamamatsu University School of Medicine)
- Kazuhiro Shiozaki (Nara Institute of Science and Technology)
- Joseph Takahashi (University of Texas, Southwestern Medical Center at Dallas, USA)
- Seiji Takashima (Osaka University Graduate School of Medicine)
- Takashi Takeuchi (Tottori University)
- Fuyuhiko Tamanoi (University of California-Los Angels, USA)
- Masashi Tanaka (Tokyo Metropolitan Institute of Gerontology)
- Jeremy Thorner (University of California, Berkeley, USA)
- Hiroki Ueda (RIKEN, Center for Developmental Biology)
- Masaru Ueno (Hiroshima University)
- Mitsuhiro Yanagida (Okinawa Institute of Science and Technology)
- Minoru Yoshida (RIKEN, Wako institute)
- Paul Young (Queen's University, Canada)Dr. Haruhiko Bito (University of Tokyo Graduate School of Medicine)
7. Other
- Prof. Mitsuhiro Yanagida received the Order of Culture on 3rd November, 2011.
- Prof. Mitsuhiro Yanagida elected a foreign associate of the National Academy of Science.



