FY2016 Annual Report
Neural Computation Unit
Professor Kenji Doya
Abstract
The Neural Computation Unit pursues the dual goals of developing robust and flexible learning algorithms and elucidating the brain’s mechanisms for robust and flexible learning. Our specific focus is on how the brain realizes reinforcement learning, in which an agent, biological or artificial, learns novel behaviors in uncertain environments by exploration and reward feedback. We combine top-down, computational approaches and bottom-up, neurobiological approaches to achieve these goals.
In FY2016, we obtained two major external grants: Kakenhi project on Artificial Intelligence and Brain Science and Post-K Supercomputing project on Brain and Artificial Intelligence.
In the Kakenhi project on Artificial Intelligence and Brain Science, Prof. Doya serves as the project leader to orchestrate research in 11 laboratories across the country, from both AI and neuroscience fields.
Prof. Doya also leads the Post-K supercomputing project on Brain and Artificial Intellignece, a joint effort of seven groups working on neural data analysis, neural network modeling, and brain-inspired AI applications to utilize Japan's next flagship supercomputer, about 100 times performance of the present K supercomputer, to launch in 2020.
Our unit also participated in Japan's major brain science project, Brain/MINDS, in developing neural data analysis pipelines and modeling methodologies.
1. Staff
Dynamical Systems Group
- Ildefons Magrans de Abril, Staff Scientist
- Carlos Enrique Gutierrez, Postdoctoral Scholar
- Hiromichi Tsukada, Postdoctoral Scholar
- Jessica Verena Schulze, OIST Student
- Kosuke Yoshida, Special Research Student
- Junichiro Yoshimoto, Visiting Researcher (NAIST)
Systems Neurobiology Group
- Makoto Ito, Group Leader
- Akihiro Funamizu, Postdoctoral Scholar
- Kazumi Kasahara, JSPS Research Fellow
- Katsuhiko Miyazaki, Staff Scientist
- Kayoko Miyazaki, Staff Scientist
- Hiroaki Hamada, OIST Student
- Tomohiko Yoshizawa, Special Research Student
Adaptive Systems Group
- Qiong Huang, OIST Student
- Farzana Rahman, OIST Student
- Chris Reinke, OIST Student
- Jiexin Wang, Special Research Student
- Paavo Parmas, OIST Student
- Tadashi Kozuno, OIST Student
- Eiji Uchibe, Visiting researcher (ATR)
Administrative Assistant / Secretary
- Emiko Asato
- Kikuko Matsuo
2. Collaborations
Development of reinforcement learning technology for large scale systems and its application to ICT systems
- Type of collaboration: Joint research
- Researchers:
- Seishi Okamoto, Fujitsu Laboratories Ltd.
- Naoki Sashida, Fujitsu Laboratories Ltd.
- Tomotake Sasaki, Fujitsu Laboratories Ltd.
3. Activities and Findings
3.1 Kakenhi Project on Artificial Intelligence and Brain Science
1) In applying deep neural networks to reinforcement learning, standard methods like Deep Q-Network (DQN) take a method to assure stability at the compromise of data-efficiency. We generalized the method in the framework of "approximate value iteration" and mathematically analyzed the speed of convergence. As a result, we derived a more data-efficient algorithm and verified its performance in benchmark tasks.
2) In order to elucidate the information coding in the basal ganglia for reinforcement learning, we performed cell-type specific optical recording of neurons in the striosome compartment of the striatum. In an odor conditioning task, we discovered that striosome neurons acquire reward predictive activities, supporting the hypothesis that they encode state value functions in reinforcement learning.
3) To understand the mechanism of predictive coding in the cerebral cortex, we started to develop a novel behavioral paradigm for mice and calcium imaging system using a prism lens for simultaneously recording neurons in multiple cortical layers.
Figure 1: Endoscopic calcium imaging of striosome neurons.
3.2 Post-K Supercomputing Project on Brain and Artificial Intelligence
We constructed an “integrate-and-fire” neural network model of the basal ganglia circuit based on the mean-field approximation model by Lineard & Girarad (2014), which was based on the anatomical and physiological data of the basal ganglia. The model was coded by a network description language PyNEST and scaled as 1/20,000 of the macaque monkey brain, with 2,644 neurons in the input site, the striatum.
In parallel simulation by NEST, we succeeded in reproducing the firing rates of each region in normal and pharmacologically manipulatesstates. Furthermore, by taking this network as a single “channel” and laterally connecting multiple channels, we observed that only a single channel produced its output for a given input, which was a realization of action selection function.
Figure 2: Spiking neural network model of the basal ganglia.
3.3 Brain/MINDS Project on Multi-scale Neural Modeling
The aim of this research and development is to maximally utilize the data obtained in the Brain/MIDNS program at macroscopic, mesoscopic and microscopic levels for building computational models of the brain and to integrate the models at different levels for understanding and predictions of cognitive and behavioral functions starting from the molecular and cellular levels through the circuit mechanisms. We performed the following research and developments:
1) Automatic estimation of model parameters: We explored the Bayesian connectivity inference framework to integrate the connections estimated by fiber-tracking using diffusion MRI data as the prior probability and the resting-state functional MRI data for the likelihood and ran benchmarks of multiple methods. We also produced data processing pipeline to apply those methods for the marmoset MRI data obtained at RIKEN.
2) Multi-scale data integration: Toward the goal of evaluation and optimization of fiber-tracking algorithms for diffusion MRI data using the neural tracer data as the reference, we built trace image processing pipeline in close collaboration with researchers at RIKEN and Kyoto University. We also performed evaluation of fiber-tracking algorithms.
3) Model building and performance validation: We built a whole-brain network model based on the connectivity matrix estimated by fiber tracking based on the marmoset diffusion MRI data and analyzed how the model could reproduce the functional connectivity given by resting-state functional MRI. We clarified key parameters that affect the network behaviors.
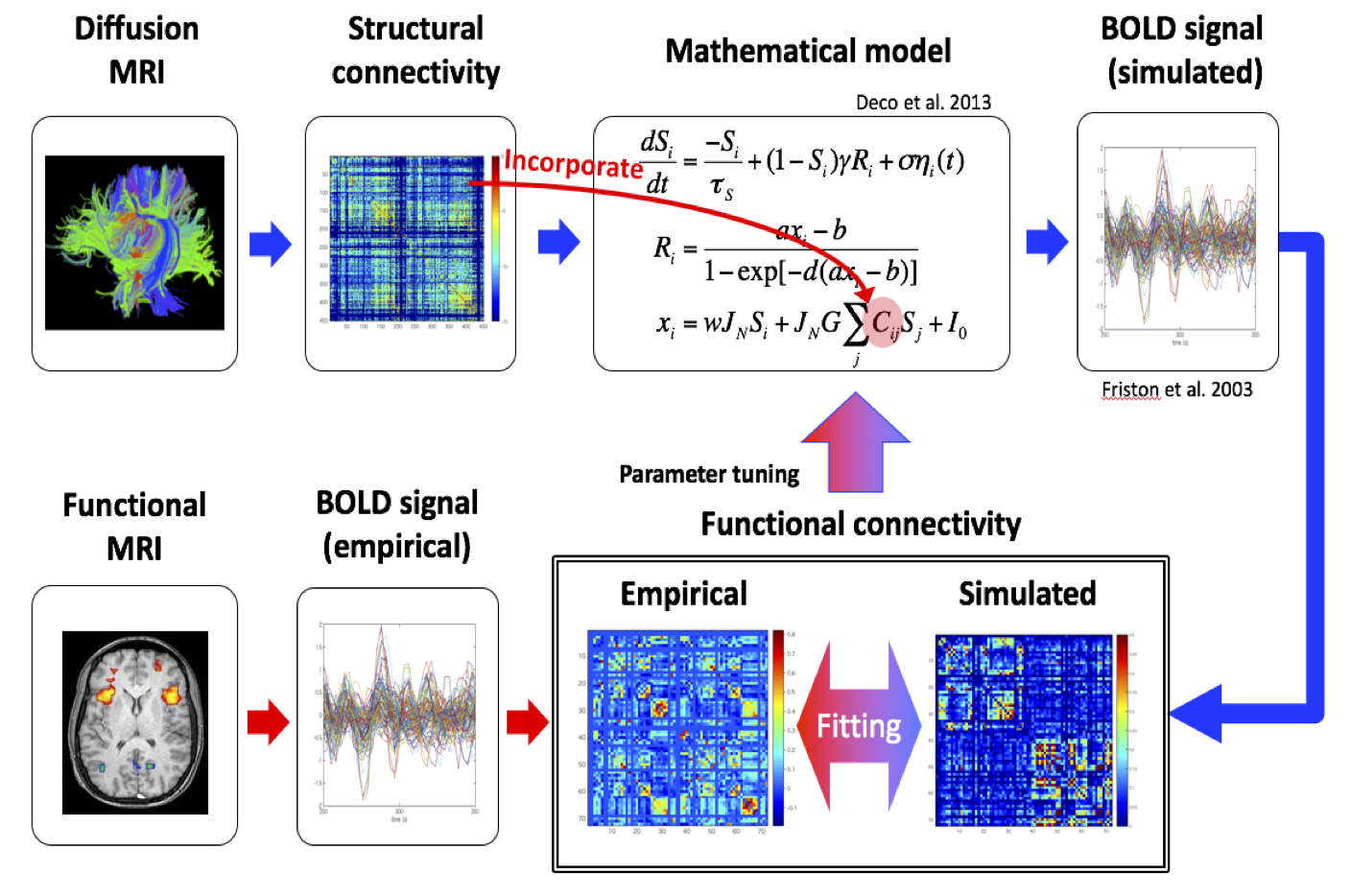
Figure 3: Simulation of network model based on anatomical connectivity data from diffusion MRI and its comparison with functional connectivity data from resting-state functional MRI.
3.4 Other Research Developments
We published papers from our previous Kakenhi project on Prediction and Decision Making, regarding the neural mechanisms of mental simulation (Fermin et al., Scientific Reports, 2016; Funamizu et al., Nature Neuroscience, 2016).
We also published papers on efficient reinforcement learning algorithms (Elfwing et al., 2016; Wang et al., 2017).
4. Publications
4.1 Journals
- Elfwing, S., Uchibe, E., & Doya, K. (2016). From free energy to expected energy: Improving energy-based value function approximation in reinforcement learning. Neural Networks, 84, 17-27. doi:http://dx.doi.org/10.1016/j.neunet.2016.07.013
- Fermin, A. S. R., Yoshida, T., Yoshimoto, J., Ito, M., Tanaka, S. C., & Doya, K. (2016). Model-based action planning involves cortico-cerebellar and basal ganglia networks. Nature Scientific Reports, 6(31378). doi:10.1038/srep31378
- Funamizu, A., Kuhn, B., & Doya, K. (2016). Neural substrate of dynamic Bayesian inference in the cerebral cortex. Nature Neuroscience. doi:http://dx.doi.org/10.1038/nn.4390
- Okamoto, Y., Okada, G., Tanaka, S., Miyazaki, K., Miyazaki, K., Doya, K., & Yamawaki, S. (2016). The role of serotonin in waiting for future rewards in depression. International Journal of Neuropsychopharmacology, 19, 33-33.
- Shimizu, Y., Doya, K., Okada, G., Okamoto, Y., Takamura, M., Yamawaki, S., & Yoshimoto, J. (2016). Depression severity and related characteristics correlate significantly with activation in brain areas selected through machine learning. International Journal of Neuropsychopharmacology, 19, 135-136.
- Shouno, O., Tachibana, Y., Nambu, A., & Doya, K. (2017). Computational model of recurrent subthalamo-pallidal circuit for generation of parkinsonian oscillations. Frontiers in Neuroanatomy, 11. doi:http://dx.doi.org/10.3389/fnana.2017.00021
- Wang, J. X., Uchibe, E., & Doya, K. (2017). Adaptive baseline enhances EM-based policy search: Validation in a view-based positioning task of a smartphone balancer. Frontiers in Neurorobotics, 11. doi:http://dx.doi.org/10.3389/fnbot.2017.00001
- Yoshida, K., Yoshimoto, J., & Doya, K. (2017). Sparse kernel canonical correlation analysis for discovery of nonlinear interactions in high-dimensional data. BMC Bioinformatics, 18(1), 108. doi:http://dx.doi.org/10.1186/s12859-017-1543-x
4.2 Books and other one-time publications
- Doya, K. (2016). Reinforcement learning as a model of intelligence. In T. Makino, T. Shibuya, & S. Shirakawa (Eds.), Reinforcement Learning From Now On (pp. 284-294). Tokyo: Morikita Publishing Co., Ltd.
4.3 Oral and Poster Presentations
- Doya, K. (2016.09.08-10). Artificial Intelligence, Brain Science and Human Mind, Waters Edge, Colombo, Sri Lanka.
- Doya, K. (2016). Brain Science and Artificial Intelligence: From Basic Science to Innovation. Naha, Okinawa: Okinawa Open Days 2016(OOD2016).
- Doya, K. (2016.11.22). Imaging the neural circuits for mental simulation and reward prediction, University of Toyama.
- Doya, K. (2016.06.21). Introduction to reinforcement learning and Bayesian inference, OIST Seaside House, Onna-son, Okinawa.
- Doya, K. (2016.9.26-27). Neural Circuit Mechanisms of Mental Simulation, Hebrew University of Jerusalem, Israel.
- Doya, K. (2017.03.29). Coding of action and state values in the striatal compartments, Merida, Mexico.
- Doya, K. (2017). From Circuit Architectures to Learning Mechanisms of the Cerebellum, the Basal Ganglia, and the Cerebral Cortex. Yokohama, Kanagawa: The64thJSAP Spring Meeting, 2017.
- Doya, K., Elfwing, S., & Uchibe, E. (2016.10.08). Design, inference and evolution of reward functions for robots, University of London, London.
- Doya, K., Miyazaki, K. W., & Miyazaki, K. (2016.09.28-30). Serotonin and the regulation of patience, Gatsby Computtional Neuroscience Unit, London, England.
- Funamizu, A. (2016). Neural substrate of dynamic Bayesian inference in posterior parietal cortex. OIST, Okinawa, Japan.
- Funamizu, A. (2016). Neural substrate of dynamic Bayesian inference in posterior parietal cortex. Pacifico Yokohama, Yokohama, Kanagawa, Japan.
- Funamizu, A. (2016). Neural substrate of dynamic Bayesian inference in the cerebral cortex. New York University, New York, US.
- Funamizu, A., Kuhn, B., & Doya, K. (2016.07.20). Neural substrate of dynamic Bayesian inference in posterior parietal cortex, Pacifico Yokohama, Yokohama, Kanagawa, Japan.
- Gutierrez, C. E., Yoshimoto, J., & Doya, K. (2016). Community Detection and Mean Field Approximation for Dimension Reduction of Spiking Network Models. Suzuki Umetaro Hall RIKEN, Wako, Saitama, Japan: Advances in Neuroinformatics (AINI) 2016.
- Hamada, H., Hikishima, K., Takata, N., Sakai, Y., Tanaka, K., & Doya, K. (2016). The difference of resting-state brain activities in awake and anesthetized states in mice. University of Vienna,Vienna, Austria: 5th Biennial Conference on Resting State and Brain Connectivity 2016.
- Hamada, H., Sakai, Y., Takata, N., Hikishima, K., Tanaka, K., & Doya, K. (2016). Mapping Functional Whole-Brain Networks in an Awake State of Mice. Pacifico Yokohama, Yokohama, Kanagawa, Japan: The 39th Annual Meeting of the Japanese Neuroscience Society.
- Ito, M. (2016). Extraction of brain macrostate and estimation of algorism by sparse modeling. Takeda Hall, Tokyo university, Tokyo, Japan: 4th research meeting of intiative for high-dimensional data-driven science through deepening of sparse modeling.
- Ito, M. (2016). Hippcampal CA1 recording and macro-state extraction. Keio University, Tokyo: Public symposium of Sparse Modeling.
- Ito, M., & Doya, K. (2016.07.22). Nonlinear dimension reduction of optically recorded activities of hippocampus CA1 pyramidal neurons revealed not only spatial but also motion information coding, Pacifico Yokohama, Yokohama, Kanagawa, Japan.
- Ito, M., & Doya, K. (2016.11.13). A nonlinear unsupervised-learning method extracts spatial information and more from hippocampal population activity of freely-moving rats San Diego, CA, USA.
- Ito, M., & Yoshizawa, T. (2016). Endoscope calcium imaging of rat hippocampus and mouse striatum by miniature microscope. 8th Optogenetics symposium. Mita campus, Keiou-Univ, Tokyo, Japan.
- Magrans de Abril, I., Yoshimoto, J., & Doya, K. (2016). A strategy to infer hidden sources behind multiple neural recording data for neural circuit inference. Suzuki Umetaro Hall RIKEN, Wako, Saitama, Japan: 4th INCF Japan Node International Workshop Advances in Neuroinformatics 2016 and 14th INCF Nodes Workshop.
- Qiong, H., Uchibe, E., & Doya, K. (2016). Emergence of communication among reinforcement learning agents under coordination environment. Cergy-Pontoise, Paris, France: IEEE ICDL-EPIROB 2016.
- Reinke, C. (2016). Brain inspired temporal decision making algorithms & Research opportunities at the Okinawa Institute of Science and Technology. Frankfurt Institute for Advanced Studies.
- Schulze, J. V. (2016). Functionally informed priors in a bayesian machine lerning approach to neural connectivity inference. the Universidad Catolica San Pablo, Arequipa, Peru: Machine learning summer school 2016
- Schulze, J. V. (2016). Functionally Infotmend Priors in a Bayesian Machine Learning Approach to Neural Connectivtity Inference. Barcelona, Spain: WiML & NIPS.
- Tsukada, H., Hamada, H., Nakae, K., Ishii, S., Hata, J., Okano, H., & Doya, K. (2016). Mathematical modeling and dynamical analysis using structural and functional connectivity. Suzuki Umetaro Hall RIKEN, Wako, Saitama, Japan: 4th INCF Japan Node International Workshop, Advances in Neuroinformatics 2016 and 14th INCF Nodes Workshop.
- Tsukada, H., Hamada, H., Nakae, K., Ishii, S., Hata, J., Okano, H., & Doya, K. (2016.08.31). A mathematical modeling approach for structural and functional connectivity in MRI, OIST Seaside House, Onna-son,Okinawa, Japan.
- Tsukada, H., Hamada, H., Nakae, K., Ishii, S., Hata, J., Okano, H., & Doya, K. (2017). Dynamical analysis and mathematical modeling using structural and functional connectivity data. OIST Seaside House,Onna, Okinawa, Japan.
- Tsukada, H., Hamada, H., Nakae, K., Ishii, S., Hata, J., Okano, H., & Doya, K. (2017). Mathematical model using structural and functional connectivity data of marmoset. Hokkaido university, Sapporo, Hokkaido, Japan.
- Tsukada, H., Hamada, H., Nakae, K., Ishii, S., Hata, J., Okano, H., & Doya, K. (2017). Whole brain model simulation based on diffusion and functional MRI data. IINO HALL, Chiyoda-KU, Tokyo: The second symposium of brain and mind.
- Uchibe, E. (2016). Deep inverse reinforcement learning based on KL-control.Special research committee on embodiment cognitive science and real world applications (ECSRA). Graduate School of Engineering Science, Osaka University, Osaka, Japan.
- Yoshimoto, J., Kannon, T., Amano, M., Nishioka, T., Usui, S., & Kaibuchi, K. (2016). KANPHOS Platform: A database for neural phosphoproteomics with quality control. Suzuki Umetaro Hall RIKEN, Wako, Saitama, Japan: The 4th INCF Japan Node International Workshop Advances in Neuroinformatics (AINI 2016).
- Yoshizawa, T., Ito, M., & Doya, K. (2016). The activities of striatal patch neurons in classical conditioning. Rusutsu, Hokkaido, Japan: The 16th Winter Workshop on the Mechanism of Brain and Mind.
- Yoshizawa, T., Ito, M., & Doya, K. (2016). The role of striatal patch neurons in reward-based learning. Pacifico Yokohama, Yokohama, Kanagawa, Japan: The 39th Annual Meeting of the Japan Neuroscience Society.
- Yoshizawa, T., Ito, M., & Doya, K. (2016). The striatal striosome compartment encodes the value of sensory stimulus. San Diego Convention Center, San Diego, CA, USA: Neuroscience 2016.
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminars
Computational model-based analysis of learning and memory: stress, genes and prediction
- Date: May 11, 2016
- Venue: OIST Campus Lab1
- Speaker: Dr. Gediminas Luksys (University of Basel)
Learning word meaning and associations from co-occurrence data
- Date: September 23, 2016
- Venue: OIST Campus Lab1
- Speaker: Dr. Stefan Evert (Friedrich-Alexander-Universität Erlangen-Nürnberg)
Is depression caused by a hyperactive habenula?
- Date: November 29, 2016
- Venue: OIST Campus Lab1
- Speaker: Dr. Jonathan Roiser (UCL Institute of Cognitive Neuroscience (ICN))
Action Selection & Reinforcement Learning in animals & robots
- Date: December 5, 2016
- Venue: OIST Campus Lab1
- Speaker: Dr. Benoit Girard (ISIR, UPMC/CNR)
Saccadic eye movements
- Date: December 6, 2016
- Venue: OIST Campus Lab1
- Speaker: Dr. Benoit Girard (ISIR, UPMC/CNR)
Navigation strategies & Multiple learning systems
- Date: December 7, 2016
- Venue: OIST Campus Lab1
- Speaker: Dr. Benoit Girard (ISIR, UPMC/CNR)
6.2 Events
Joint Workshop: Neuro-Computing, Bioinformatics, Mathematical modeling and Machine Learning
- Date: July 4-6, 2016
- Venue: OIST Capmpus, Semiar Room B250 & Seminar Room C210
- Co-organizers:
- The Institute of ElectronicsInformation and Communication Engineers(IEICE)
- Information Processing Society of Japan
- IEEE Computational Intelligence Society Japan Chapter
- Japan Neural Network Society
- Speaker:
- Dr. Yutaka Hirata (Chubu University)
Sponsored Research Meeting: "Comparison and Fusion of Artificial Intelligence and Brain Science" and "Next Generation of Kei Super Computer" joint workshop
- Date: August 11-12, 2016
- Venue: Shonan Village Center
- Organizers:
- Grant-in Aid for Scientific Research on Innovative Areas, MEXT, JAPAN, Correspondence and Fusion of Artificial Intelligence and Brain Science
- Post K Exploratory Challenge, Big Brain-Data Analysis, Whole Brain-Scale Simulation, and Brain -Style AI Architecture
- Speaker:
- Dr. Kenji Doya (OIST)
- Dr. Shin Ishii (Kyoto University)
- Dr. Masashi Sugiyama (University of Tokyo)
- Dr. Yutaka Matsuo (University of Tokyo)
- Dr. Masamichi Sakagami (Tamagawa University)
- Dr. Jun Igarashi (RIKEN)
- Dr. Tadashi Yamazaki (University of Electro-Communications)
- Dr. Koichi Takahashi (RIKEN)
- Dr. Hiroshi Yamakawa (Dowango)
- Dr. Tatsuya Harada (University of Tokyo)
Sponsored Research Meeting: The 1st Research Area Meeting Scientific Research on Innovative Areas: Artificial Intelligence and Brain Science
- Date: December 21, 2016
- Venue: National Center of Science
- Organizer: Grant-in Aid for Scientific Research on Innovative Areas, MEXT, JAPAN, Correspondence and Fusion of Artificial Intelligence and Brain Science



