FY2020 Annual Report
Neural Coding and Brain Computing Unit
Professor Tomoki Fukai

Abstract
Our goal is to construct a minimal but yet effective description of powerful and flexible computation implemented by the brain’s neural circuits. To achieve this goal, we attempt to uncover the neural code and circuit mechanisms of brain computing through computational methodologies and neural data analyses. This year, we constructed a microcircuit model to explore the mechanisms of memory-related cross-area communications. We also explored the neural representations of salient features in reward-driven spatial navigation.
1. Staff
- Staff Scientist
- Chi Chung Fung, PhD
- Tatsuya Haga, PhD
- Postdoctoral Scholar
- Ibrahim Alsolami, PhD
- Hongjie Bi, PhD
- Toshitake Asabuki, PhD
- Research Unit Technician
- Ruxandra Cojocaru, PhD
- PhD Student
- Thomas Burns
- Gaston Sivori
- Maria Astrakhan
- Roman Koshkin
- Munenori Takaku
- Special Research Student
- Milena Menezes Carvalho
- Rotation Student
- Takaku Munenori (Term 3)
- Jose Carlos Pelayo (Term 3)
- Natalya Weber (Term 1)
- Morihiro Ohta (Term 2)
- Hideyuki Yoshimura (Term 2)
- Research Intern
- Hugo Musset (Feb-Jul 2020)
- Meile Petrauskaite (Oct 2020-Mar 2021)
- Research Unit Administrator
- Kiyoko Yamada
2. Collaborations
2.1 "Idling Brain" project
kaken.nii.ac.jp/en/grant/KAKENHI-PROJECT-18H05213
- Supported by Grant-in-Aid for Specially Promoted Research (KAKENHI)
- Type of collaboration: Joint research
- Researchers:
- Professor Kaoru Inokuchi, University of Toyama
- Dr. Khaled Ghandour, University of Toyama
- Dr. Noriaki Ohkawa, University of Toyama
- Dr. Masanori Nomoto, University of Toyama
2.2 Chunking sensorimotor sequences in human EEG data
- Type of collaboration: Joint research
- Researchers:
- Professor Keiichi Kitajo, National Institute for Physiological Sciences
- Dr. Yoshiyuki Kashiwase, OMRON Co. Ltd.
- Dr. Shunsuke Takagi, OMRON Co. Ltd.
- Dr. Masanori Hashikaze, OMRON Co. Ltd.
2.3 Cell assembly analysis in the songbird brain
- Type of collaboration: Internal joint research
- Researchers:
- Professor Yoko Yazaki-Sugiyama, OIST
- Dr. Jelena Katic, OIST
2.4 A novel mathematical approach to partially observable Markov decision processes
- Type of collaboration: Joint research
- Researchers:
- Haruki Takahashi, PhD student, Kogakuin University
- Prof. Takashi Takekawa, Kogakuin University
- Prof. Yutaka Sakai, Tamagawa University
3. Activities and Findings
3.1 Oscillatory memory processing in an Entorhinal-Hippocampal microcircuit model
Task-relevant information is processed by multiple brain regions. In this modeling study, we explored how this multiarea computation is conducted in the entorhinal-hippocampal local circuits during spatial working memory tasks (Fig. 1). In the model, theta oscillation coordinates the proper timing of interactions between the two regions. The model predicts the concentration of acetylcholine (ACh) is increased from the default level (low [Ach]) during the encoding (high [Ach]), maintenance (modestly high [Ach]), and retrieval (high [Ach]) of spatial working memory. Our model also predicts that the medial entorhinal cortex (MEC) is engaged in decoding as well as encoding spatial memory, which we confirmed by experimental data analysis. This work appeared in Kurikawa et al. Cerebral Cortex (2020).
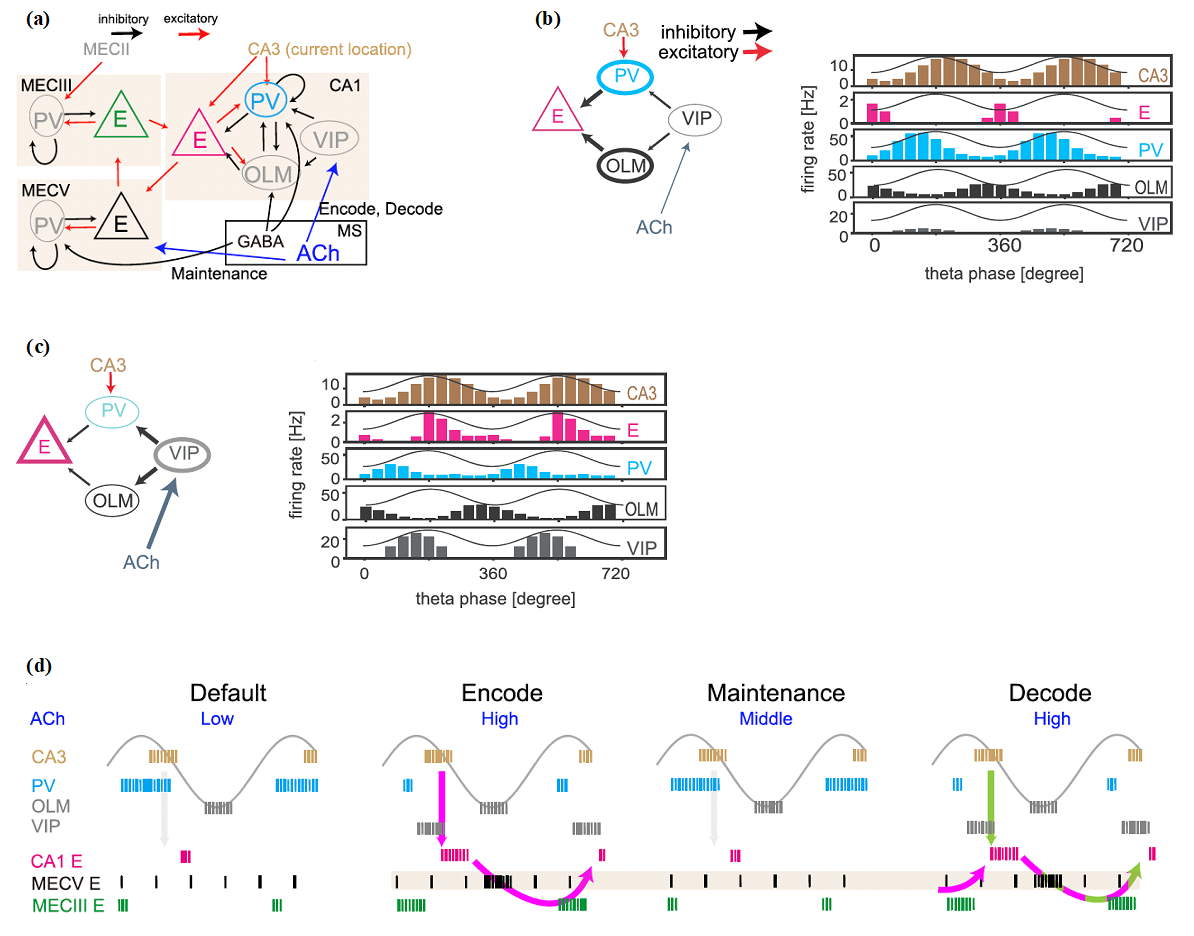
Figure 1: Cross-area communications in the entorhinal-hippocampal microcircuit model. (a) The connectivity of the network model is shown. Acetylcholine (ACh) regulates vasoactive intestinal polypeptide (VIP) neurons to control CA1 excitatory cells during encoding and decoding epochs and modulate the sensitivity of excitatory cells in the layer-5 medial entorhinal cortex (MECV) during encoding, maintenance, and decoding epochs. (b) In the default epoch in which [ACh] is low, output from VIP neurons is weakened and, consequently, parvalbumin (PV) and oriens-lacunosum moleculare (OLM) neurons are strongly activated to suppress the CA3-to-CA1 signaling around the peaks and the troughs of theta oscillation, respectively. Accordingly, CA1 excitatory neurons rarely fire around the peaks and troughs of theta oscillation. (c) In contrast, the gate of the CA3-to-CA1 signaling is opened during the epochs of high [ACh]. Now, CA1 excitatory neurons show strong activation immediately after the peaks of theta oscillation because PV and OLM neurons are suppressed around the peaks. (d) The resultant memory processing is schematically illustrated. In the default epoch (before encoding), [ACh] is low and the activity of PV is high enough to inhibit the activity of CA1 excitatory neurons and the flow of spatial information from CA3 to CA1. In the encoding epoch,[ACh] is set higher and VIP neurons are activated, which in turn inhibits CA1 PV neurons to enable the transfer of spatial information into CA1. Accordingly, the spatial information is stored in the CA1–MECV–MECIII loop circuit (magenta arrows in the panel of encoding epoch). In the maintenance epoch, [ACh] is at a middle level and PV neuron activity is again high, blocking the reentry of location information into CA1. During this blockade, the spatial information is maintained by the activation of calcium-dependent cation current in MECV excitatory neurons without spike generation. Finally, in the decoding epoch, [ACh] is high again and spatial information is loaded from CA3 to CA1 (green arrow). It is noted that information on the previous location maintained in MECV is also reloaded from MECIII to CA1. Therefore, CA1 exhibits both current and previous position-encoding activities during the decoding epoch.
3.2 Differential representations of landmarks and rewards in the hippocampus
In this collaboration, we used a virtual reality system to explore how the locations associated with salient features such as landmarks and rewards are represented in the hippocampus. We found a disproportionately large number of neurons representing such locations (Fig. 2). Interestingly, in mice lacking Shank2, an autism spectrum disorder (ASD)-linked gene encoding an excitatory postsynaptic scaffold protein, the learning-induced over-representation of landmarks was absent whereas that of rewards was substantially increased. Our findings demonstrate that the hippocampal representations of different salient features undergo different coding processes in the generation of hippocampal saliency maps. The result was published in Sato et al., Cell Report (2020).

Figure 2: Representations of landmarks and rewards in CA1. (a) Visual information including landmarks and reward points was given to mice running on a trackball through a virtual reality system. (b) As learning proceeded, the hippocampus recruited an increased number of place cells (PCs) for representing the locations of landmarks and rewards.

3.2 Preprints under review
-
Tatsuya Haga and Tomoki Fukai. Associative memory networks for graph-based abstraction. bioRxiv 2020.12.24.424371, doi.org/10.1101/2020.12.24.424371.
In this paper, we propose an extended attractor network model for the graph-based hierarchical computation, called Laplacian associative memory. This model generates multiscale representations for communities (clusters) of associative links between memory items and connects graph theory and the attractor dynamics to provide a biologically plausible mechanism for abstraction in the brain.
-
Toshitake Asabuki and Tomoki Fukai. Neural mechanisms of context-dependent segmentation tested on large-scale recording data. bioRxiv2021.04.25.441363, doi.org/10.1101/2021.04.25.441363.
We show that a recurrent gated network of neurons with dendrites can context-dependently solve difficult segmentation tasks. Dendrites in this model learn to predict somatic responses in a self-supervising manner, as shown previously (Asabuki and Fukai, 2020) while recurrent connections learn a context-dependent gating of dendro-somatic current flows to minimize a prediction error. The model shows excellent performance in segmenting cell assembly patterns in both synthetic sequence data and large-scale calcium imaging data.
4. Publications
4.1 Journals
- Tomoki Kurikawa, Kenji Mizuseki, Tomoki Fukai (2020) Oscillation-Driven Memory Encoding, Maintenance, and Recall in an Entorhinal–Hippocampal Circuit Model, Cerebral Cortex, bhaa343, DOI: https://doi.org/10.1093/cercor/bhaa343
- Masaaki Sato, Kotaro Mizuta, Tanvir Islam, Masako Kawano, Yukiko Sekine, Takashi Takekawa, Daniel Gomez-Dominguez, Alexander Schmidt, Fred Wolf, Karam Kim, Hiroshi Yamakawa, Masamichi Ohkura, Min Goo Lee, Tomoki Fukai, Junichi Nakai and Yasunori Hayashi (2020) Distinct mechanisms of over-representation of landmarks and rewards in the hippocampus. Cell Rep, 32, 107864:1-14, DOI: https://doi.org/10.1016/j.celrep.2020.107864
4.2 Books and other one-time publications
- Tomoki Fukai (2020) 細胞体−樹状突起間の整合性確認による時間的特徴の分割 (Japanese). KAKENHI Project (AI & Brain Science) News Letter, vol. 8: 12-13. http://www.brain-ai.jp/wp-content/uploads/2020/10/Newsletter08.pdf
4.3 Oral and Poster Presentations
Oral Presentations
- Tomoki Fukai, Multi-faceted analysis of cell assembly code, Calcium Imaging Workshop Organized by “Correspondence and Fusion of Artificial Intelligence and Brain Science”, Online (March 2021)
- Tomoki Fukai, Creation of novel paradigms to integrate neural network learning with dendrites, The 10th Research Area Meeting Scientific Research on Innovative Areas: Artificial Intelligence and Brain Science, Online (March 2021)
- Tomoki Fukai, Rate and temporal coding perspectives of motor processing in cortical microcircuits, Online Workshop Series Neural Control: From data to machines, Online (November 2020)
- Tomoki Fukai, 情報圧縮:脳的アプローチ, 第12回数理モデリング研究会 数理モデリングの哲学--数理モデルは何のためにあるのか --, Online (November 2020)
- Tomoki Fukai, Creation of novel paradigms to integrate neural network learning with dendrites, The 9th Research Area Meeting Scientific Research on Innovative Areas: Artificial Intelligence and Brain Science, Online (October 2020)
- Tomoki Fukai, Creation of novel paradigms to integrate neural network learning with dendrites, The 8th Research Area Meeting Scientific Research on Innovative Areas: Artificial Intelligence and Brain Science, Online (June 2020)
Poster Presentations
- Milena Menezes Carvalho, Tomoki Fukai, Self-supervision mechanism of multiple dendritic compartments for temporal feature learning, 29th Annual Computational Neuroscience Meeting (CNS 2020), Online (July 2020)
5. Intellectual Property Rights and Other Specific Achievements
5.1 Grants
Kakenhi Grant-in-Aid for Scientific Research on Innovative Areas (Research in a proposed research area)
- Titile: Creation of novel paradigms to integrate neural network learning with dendrites
- Period: FY2019-2020
- Principal Investigator: Prof. Tomoki Fukai
Kakenhi Grant-in-Aid for Specially Promoted Research
- Titile: Mechanisms underlying information processing in idling brain
- Period: FY2018-2022
- Principal Investigator: Prof. Kaoru Inokuchi (University of Toyama)
- Co-Investigators: Prof. Tomoki Fukai, Prof. Keizo Takao (University of Toyama)
Kakenhi Graint-in-Aid for Early-Career Scientists
- Titile: A Comprehensive Theoretical Survey on Functionalities and Properties of Adult Neurogenesis under the Influence of Slow Oscillations
- Period: FY2019-2022
- Principal Investigator: Dr. Chi Chung Fung
5.2 Patents
Joint patent application (20-0018KG)
- Title of invention:"On-line reinforcement learning algorithms for simultaneous hidden-state sampling and reward maximization
- Inventors: Haruki Takahashi (Kogakuin Univ), Takashi Takekawa (Kogakuin Univ), Yutaka Sakai (Tamagawa Univ), Tomoki Fukai (OIST)
6. Meetings and Events
Nothing to report.
7. Other
Program Committee, Annual Computational Neuroscience Meeting (CNS2020)



