Research
 Our research is completely theoretical and uses computational methods to model the biochemical and electrophysiological behavior of neurons and microcircuits and motor control of limb movements. We also perform analysis of experimental data provided by external collaborators.
Our research is completely theoretical and uses computational methods to model the biochemical and electrophysiological behavior of neurons and microcircuits and motor control of limb movements. We also perform analysis of experimental data provided by external collaborators.
All our research is supported by in house development of dedicated software.
Molecular Modeling
![]() We use molecular modeling methods to study how signaling pathways and cell biology processes involved with synaptic plasticity are influenced by the detailed morphology of neurons and by the stochastic behavior of the reactions due to the small number of molecules. To support this work we develop a simulator program called STEPS (1) that implements the spatial Gillespie SSA in a tetrahedral grid and is meant to simulate molecular events in spines or small parts of a neuronal dendrite or axon. An extension of STEPS allows for calculation of the membrane potential on the same tetrahedral mesh so that voltage-gated ion channels can also be simulated. Recently we have succeeded in effectively parallelizing the spatial SSA (2) and STEPS (3), this allows us to simulate a complete neuron or astrocyte at the nanoscale. Another version of STEPS supports spatial modeling of vesicles.
We use molecular modeling methods to study how signaling pathways and cell biology processes involved with synaptic plasticity are influenced by the detailed morphology of neurons and by the stochastic behavior of the reactions due to the small number of molecules. To support this work we develop a simulator program called STEPS (1) that implements the spatial Gillespie SSA in a tetrahedral grid and is meant to simulate molecular events in spines or small parts of a neuronal dendrite or axon. An extension of STEPS allows for calculation of the membrane potential on the same tetrahedral mesh so that voltage-gated ion channels can also be simulated. Recently we have succeeded in effectively parallelizing the spatial SSA (2) and STEPS (3), this allows us to simulate a complete neuron or astrocyte at the nanoscale. Another version of STEPS supports spatial modeling of vesicles.
We have been using STEPS to study anomalous diffusion in spiny dendrites (4) and the stochasticity of induction of cerebellar long-term depression in Purkinje cells (5). More recently we have developed an integrated molecular model of cerebellar long-term depression and long-term potentiation (6) and are currently converting this into a spatial model that includes a detailed representation of the endosomal cycle. We are using the vesicle modeling feature to simulate neurotransmitter release from a presynaptic terminal in unprecedented detail.
Cellular Modeling and Data Analysis
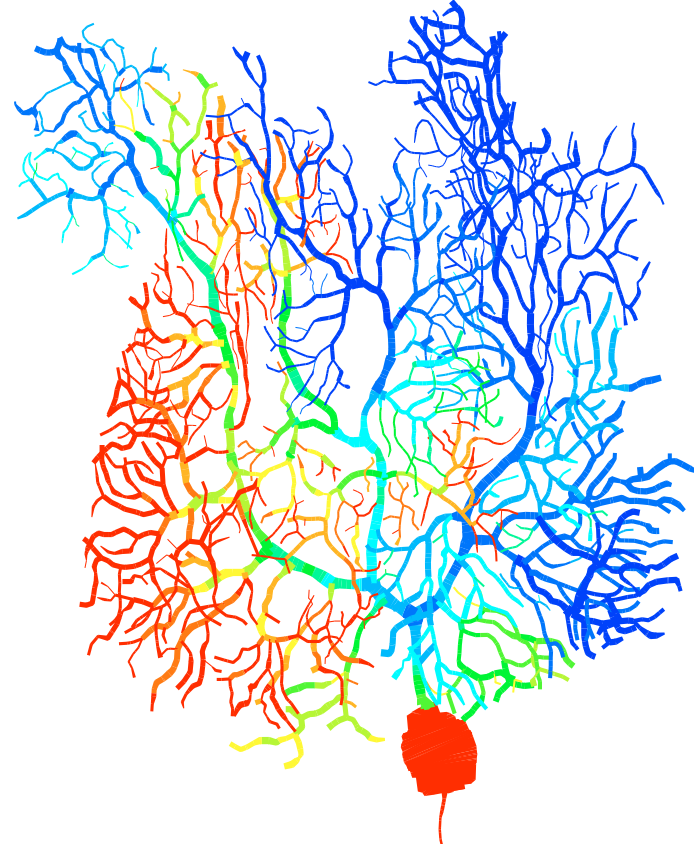
![]() Erik De Schutter is well known for the detailed model of the Purkinje cell he developed in the past (7). This model replicates the complete morphology and electrophysiology of the neuron and has demonstrated strong predictive power (8). We have now developed a new Purkinje cell model that incorporates recent data about voltage-gated channels and use it to investigate the complex spike (9). This model replicates the firing-rate dependence of the Purkinje cell Phase Response Curve, enabling firing-rate dependent oscillations (9). We also contribute to the development of models of other neurons in the cerebellum (10, 11).
Erik De Schutter is well known for the detailed model of the Purkinje cell he developed in the past (7). This model replicates the complete morphology and electrophysiology of the neuron and has demonstrated strong predictive power (8). We have now developed a new Purkinje cell model that incorporates recent data about voltage-gated channels and use it to investigate the complex spike (9). This model replicates the firing-rate dependence of the Purkinje cell Phase Response Curve, enabling firing-rate dependent oscillations (9). We also contribute to the development of models of other neurons in the cerebellum (10, 11).
We have extended our stochastic modeling to the cellular level and using the STEPS simulator (1) we demonstrated that the variability of dendritic calcium spikes, which has been observed experimentally, is caused by stochastic calcium mechanisms (12). This work links stochasticity at the molecular level with cellular properties. We are now extending this model to simulating a full Purkinje cell at the nanoscale level (13). The importance of morphological properties of astrocyte branches on calcium signaling was simulated (24).
More in general we are interested in the importance of neuronal morphology and excitability for function. We showed that the type of excitability a neuron expresses determines its type of network correlation (14, 15), an important correction of the literature on the subject.
We are extending analysis of morphology to the network level: what are the properties of the forest of dendritic trees? This is a step towards modeling the development of neuronal morphology using environmental clues. To support such modeling we are developing NeuroDevSim software, a more perfomant successor to NeuroMac (16). We are using this to model development of cerebellar cortex.
Network Modeling and Data Analysis
 We have a strong interest in cerebellar oscillations (17) which we continue to investigate in detailed 2D and 3D network models of cerebellar cortex (18). These network models comprise simplified models that capture the excitability of the neurons and detailed connectivity based on real anatomy. They are also used to investigate population coding and learning by the cerebellum and its interaction with other brain structures involved in motor control under normal conditions and in disease (19).
We have a strong interest in cerebellar oscillations (17) which we continue to investigate in detailed 2D and 3D network models of cerebellar cortex (18). These network models comprise simplified models that capture the excitability of the neurons and detailed connectivity based on real anatomy. They are also used to investigate population coding and learning by the cerebellum and its interaction with other brain structures involved in motor control under normal conditions and in disease (19).
The output of these network simulations are spike trains and we like to compare these to recordings of simple and complex spikes from Purkinje cells in rodents (20) and monkeys, which we analyze using sophisticated statistical methods. This has resulted in the discovery of a relationship between the duration of complex spikes in Purkinje cells and their firing rates (21) and of multiplexed coding (15) by simple spikes (22). Such multiplexed coding turns out to be also a property of our new Purkinje cell model (25).
References
- I. Hepburn, W. Chen and E. De Schutter: Accurate reaction-diffusion operator splitting on tetrahedral meshes for parallel stochastic molecular simulations. Journal of Chemical Physics 145: 054118 (2016).

- W. Chen and E. De Schutter: Parallel STEPS: Large Scale Stochastic Spatial Reaction-Diffusion Simulation with High Performance Computers. Frontiers in Neuroinformatics 11: 13 (2017).
- F. Santamaria, S. Wils, E. De Schutter and G.J. Augustine: Anomalous diffusion in Purkinje cell dendrites caused by dendritic spines. Neuron 52: 635-648 (2006).
- G. Antunes and E. De Schutter: A stochastic signaling network mediates the probabilistic induction of cerebellar long-term depression. Journal of Neuroscience: 32: 9288 –9300 (2012).

- A.R. Gallimore, T. Kim, K. Tanaka-Yamamoto and E. De Schutter: Switching on depression and potentiation in the cerebellum. Cell Reports 22: 722-733 (2018).

- E. De Schutter and J.M. Bower: An active membrane model of the cerebellar Purkinje cell. I. Simulation of current clamps in slice. Journal of Neurophysiology 71: 375-400 (1994).

- V. Steuber, W. Mittmann, F.E. Hoebeek, R.A. Silver, C.I. De Zeeuw, M. Häusser and E. De Schutter: Cerebellar LTD and pattern recognition by Purkinje cells. Neuron 54: 121–136 (2007).

- Y. Zang, S. Dieudonné and E. De Schutter: Voltage- and Branch-specific Climbing Fiber Responses in Purkinje Cells. Cell Reports 24: 1536–1549 (2018). Y. Zang, S. Hong and E. De Schutter: Firing rate-dependent phase responses of Purkinje cells support transient oscillations. eLife 9: 60692 (2020).
- S. Solinas, L. Forti, E. Cesana, J. Mapelli and E. De Schutter, E. D’Angelo: Computational reconstruction of pacemaking and intrinsic electroresponsiveness in cerebellar Golgi cells. Frontiers in Cellular Neuroscience 1: 2 (2007).

- V. Steuber, N. Schultheiss, A. Silver, E. De Schutter and D. Jaeger: Determinants of synaptic integration and heterogeneity in rebound firing explored with date-driven models of deep cerebellar nuclei cells. Journal of Computational Neuroscience 30: 633-658 (2011).

- H. Anwar*, I Hepburn*, H. Nedelescu, W. Chen and E. De Schutter: Stochastic calcium mechanisms cause dendritic calcium spike variability. Journal of Neuroscience: 33: 15848-15867 (2013).

- W. Chen and E. De Schutter: Time to bring single neuron modeling into 3D. Neuroinformatics 15: 1-3 (2017)
- S. Hong, S. Ratté, S. Prescott and E. De Schutter:Single neuron firing properties impact correlation-based population coding. Journal of Neuroscience 32:1413–1428 (2012).
- S. Hong, E. De Schutter and S.A. Prescott: Impact of neuronal properties on network coding: Roles of spike initiation dynamics and robust synchrony transfer. Neuron 78: 758-772 (2013).
- B. Torben-Nielsen and E. De Schutter: Context-aware modeling of neuronal morphologies. Frontiers in Neuroanatomy 8: 92 (2014).

- R. Maex and E. De Schutter: Resonant synchronization in heterogeneous networks of inhibitory neurons. Journal of Neuroscience 23: 10503-10514 (2003).

- S.K. Sudhakar, S. Hong, I. Raikov, R. Publio, C. Lang, T. Close, D. Guo, M. Negrello and E. De Schutter: Spatiotemporal network coding of physiological mossy fiber inputs by the cerebellar granular layer. PLoS Computational Biology 13: e1005754 (2017).

- P. Botta, F. M. S. de Souza, T. Sangrey, E. De Schutter and F. Valenzuela: Alcohol excites cerebellar Golgi cells by inhibiting the Na+/K+-ATPase. Neuropsychopharmacology 35: 1984-1996 (2010).

- S.-L. Shin, F.E. Hoebeek, M. Schonewille, C.I. De Zeeuw, A. Aertsen and E. De Schutter: Regular temporal patterns in cerebellar Purkinje cell simple spike trains. PLoS One 2: e485 (2007).
- P. Warnaar, J. Couto, M. Negrello, M. Junker, A. Smilgin, A. Ignashchenkova, M. Giugliano, P. Thier and E. De Schutter: Duration of Purkinje cell complex spikes increases with their firing frequency. Frontiers in Cellular Neuroscience 9: 122.
- S. Hong, M. Negrello, M. Junker, A. Smilgin, P. Thier and E. De Schutter: Multiplexed coding by cerebellar Purkinje neurons. eLife 5: e13810 (2016).
- S.O. Verduzco Flores and E. De Schutter: Draculab: A Python simulator for firing rate neural networks with delayed adaptive connections. Frontiers in Neuroinformatics 13:18 (2019).
- A. Denizot, M. Arizono, U.V. Nägerl, H. Berry and E. De Schutter: Astrocyte nanoscale morphology controls Ca2+ signals at tripartite synapses. BioRxiv Preprint (2021) https://doi.org/10.1101/2021.02.24.432635.
- Y. Zang and E. De Schutter: The Cellular Electrophysiological Properties Underlying Multiplexed Coding in Purkinje Cells. Journal of Neuroscience 41: 1850-1863 (2021).



