FY2022 Annual Report
Biological Complexity Unit
Professor Simone Pigolotti

Abstract
The Biological Complexity Unit at OIST studies the stochastic dynamics of biological and biophysical systems. Our main research lines in biophysics are the understanding of non-equilibrium mechanism of error correction in biology and modeling extended microbial populations at individual-based level. A third research line is in statistical physics and in particular in stochastic thermodynamics. FY2022 has been the fifth year of activity of our Unit. One main activity during this fiscal year has been to prepare for our Unit Review, that took place in August 2022 with a positive outcome. In addition, we have finalized projects initiated during our first five years and started new projects and collaborations.
1. Staff
- Prof. Simone Pigolotti, Associate Professor
- Dr. Robert Ross, OIST Interdisciplinary Postdoctoral Fellow
- Dr. Shrabani Mondal, Postdoctoral Scholar
- Dr. Florian Pflug, Postdoctoral Scholar
- Ms. Anzhelika Koldaeva, PhD Student
- Mr. Qiao Lu, PhD Student
- Mr. Samuel Cyrus Cure, PhD Student
- Mr. Shiling Liang, JSPS visiting Ph.D Student.
- Ms. Kaori Yamashiro, Research Unit Administrator
2. Collaborations
2.1 Speed variations of bacterial replisomes
- Description: We are collaborating with the Nucleic Acid Chemistry and Engineering Unit at OIST to understand the dynamics of bacterial replisomes theoretical and experimental perspective.
- Type of collaboration: Joint research
- Researchers:
- Yohei Yokobayashi (OIST)
- Samuel Hauf (OIST)
- Charles Plessy (OIST)
2.2 Bacterial growth in microchannels
- Description: We are collaborating with the Micro/Bio/Nanofluidics Unit at OIST to understand bacterial growth in microchannel from a theoretical and experimental perspective.
- Type of collaboration: Joint research
- Researchers:
- Amy Shen (OIST)
- Hsie-Fu Tsai (OIST)
2.3 Cellular energetics
- Description: We are continuing an international collaboration to understand the energy consumption and dissipation of cells using non-equilibrium thermodynamics.
- Type of collaboration: Joint research
- Researchers:
- Pablo Sartori (Gulbekian Institute, Lisbon, Portugal)
- Jonathan Rodenfels (MPI-CBG, Dresden, Germany).
2.4 Error correction in PCR
- Description: We are collaborating with researchers at Tohoku university to understand accuracy of Polymerase Chain Reaction by combining experiments and theory based on non-equilibrium statistical physics.
- Type of collaboration: Joint research
- Researchers:
- Shoichi Toyabe (Tohoku University)
- Hiroyuki Aoyanagi (Tohoku University).
2.5 Martingales in physics
- Description: We have been collaborating with international researchers for a review on the application of the concept of Martingales in theoretical physics
- Type of collaboration: Joint research
- Researchers:
- Edgar Roldan (ICTP Italy)
- Izaak Neri (King's College London)
- Raphael Chetrite (CNRS Nice)
- Shamik Gupta (TATA Institute Mumbai)
- Frank Julicher (MPIPKS Dresden)
- Ken Sekimoto (ESPCI Paris)
3. Activities and Findings
3.1 Speed variation of bacterial replisomes.
In living cells, DNA replication is carried out by large protein complexes called replisomes. The DNA replication program in bacteria has been qualitatively studied by looking at the abundance of DNA fragments in an exponentially growing population. We developed a quantitative theory to extract biophysically relevant information from these patterns. Our approach revealed regular, reproducible oscillations in the replisome speed in E.coli. These oscillations are correlated with previously measured variations of the mutation rate, suggesting that these patterns are related by a speed-error tradeoff. We discussed possible origins of this phenomenon.
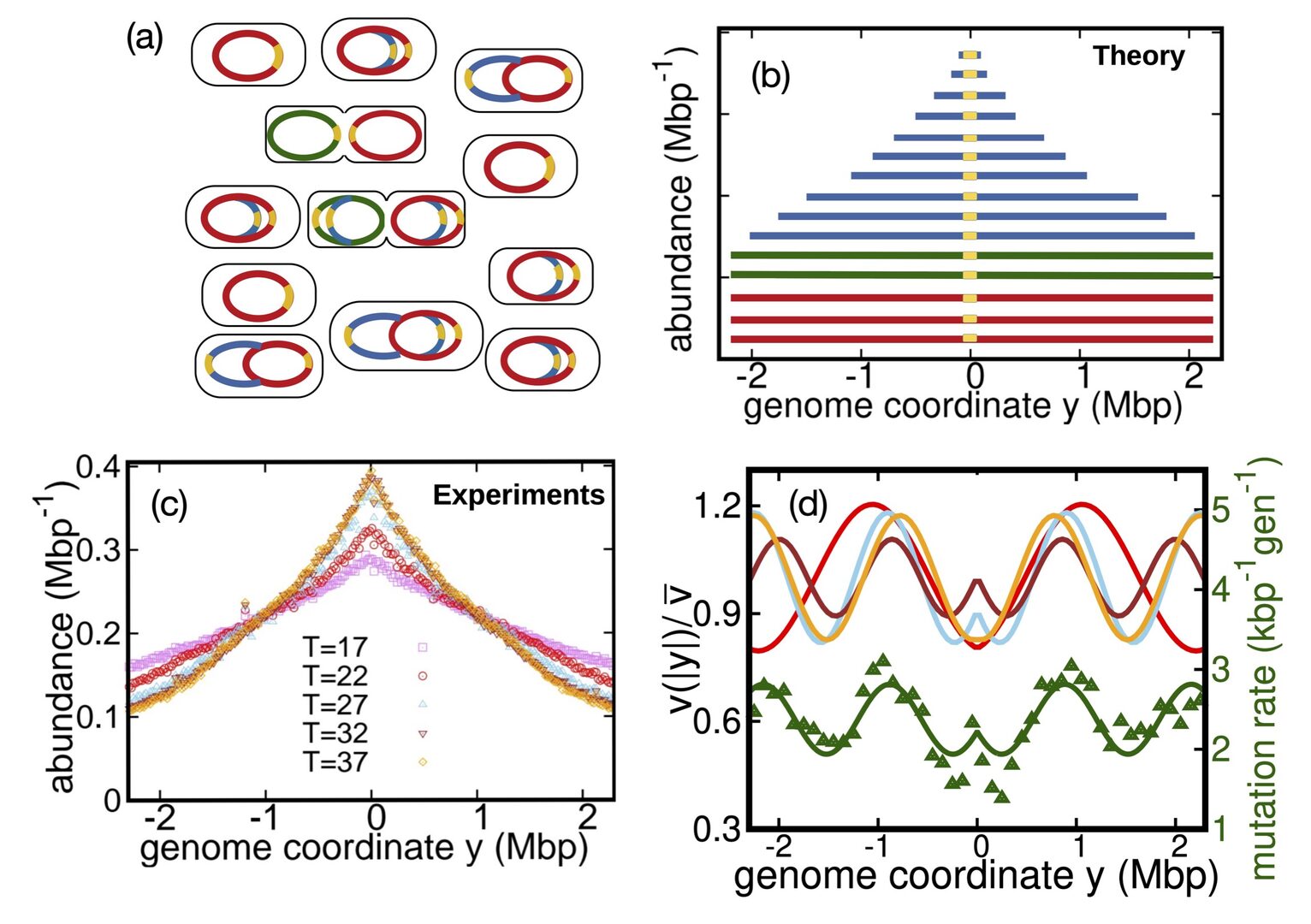
Figure 1: Inferring speed variations of bacterial replisomes. (a) An exponentially growing population of bacteria, with cells at different stages of their cell cycle. (b) DNA abundance distribution predicted by the model. (c) Experimentally measured DNA abundance in E.coli grown at different temperatures. (d) Speed variations inferred by the model, compared with variations of the previously measured local mutation rate (from Niccum et al. 2018).
3.2 Pareto optimal front of kinetic proofreading
Biological information encoded in heteropolymers needs to be replicated at high speed and with a high accuracy, i.e. a low error rate. Thermodynamic analyses of these processes show that the performance of these machines must come at a cost in terms of energy dissipation. However, this tradeoffs are difficult to characterize theoretically. We developed a numerical method based on mathematical optimization theory to compute the optimal solutions in terms of speed, error, and dissipation for the Hopfield model, a paradigmatic example of biological error correction. Our simulations reveal how the optimal solutions depend on the number of proofreading steps characterizing the reaction network.
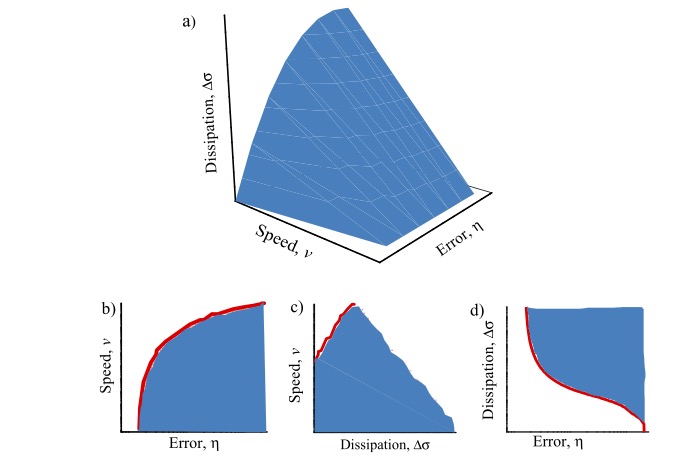
Figure 2: (a) Error-speed-dissipation optimal Pareto front for the kinetic proofreading model. (b) Error-speed Pareto front. (c) Dissipation-speed Pareto front. (d) Error-dissipation Pareto front.
3.3 Coalescent dynamics of planktonic communities
Predicting the biodiversity of planktonic community at the individual level is computationally very hard. In our previous work, we proposed a model based on coalescent theory that can efficiently simulate very large communities. We have recently shown that our coalescent model, under certain conditions, is dual to a population model including birth, density-dependent death, and immigration/speciation events. We used the model to predict the impact of turbulence on the biodiversity of a marine community, and compared with experimental results from metabarcoding studies.
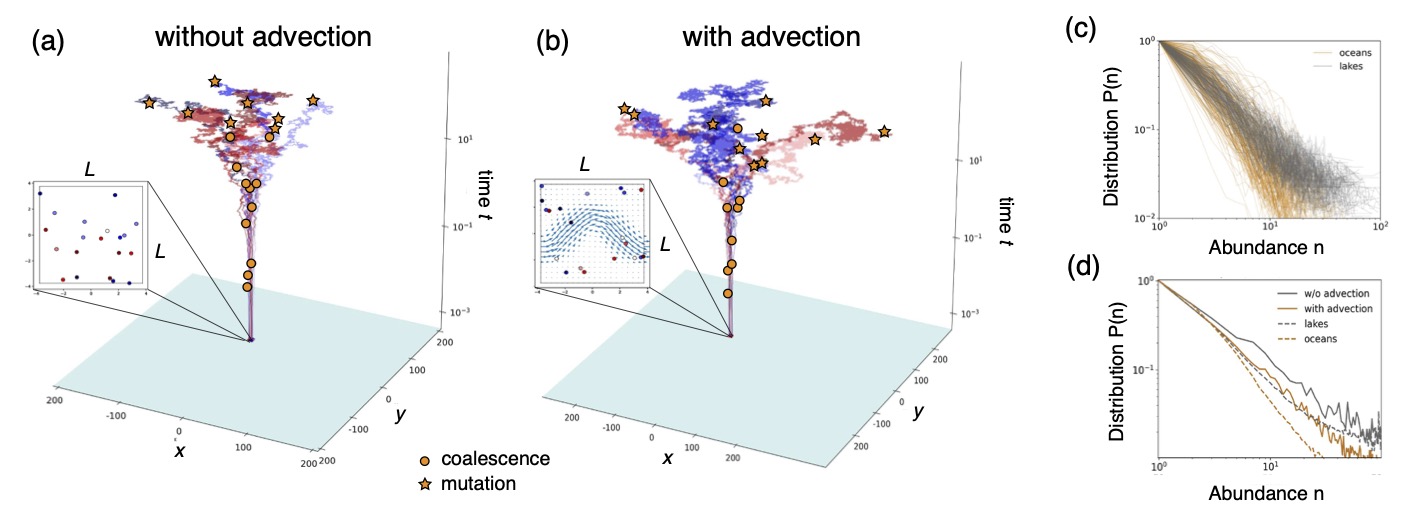
Figure 3: (a) Coalescent dynamics in the absence of advection. (b) Coalescent dynamics in the presence of advection. (c) OTU-abundance curves in oceans and lakes, as measured in metabarcoding studies (d) OTU-abundance curves predicted by the model in the presence and absence of advection.
4. Publications
4.1 Journals
- D. Chiuchiù, S. Mondal, S. Pigolotti, "Pareto optimal front of kinetic proofreading", arXiv/2208.09381 , New Journal of Physics 25, 043007 (2023).
- H. Aoyanagi, S. Pigolotti, S. Ono. S. Toyabe, "Error-suppression mechanism of PCR by blocker strands", bioRxiv preprint , Biophys. Jour. 122(7), 1334-1341 (2023).
- P.V. Martín, A. Koldaeva, S. Pigolotti, "Coalescent dynamics of planktonic communities", Phys. Rev. E 106, 044408, arXiv/2204.04609, (2022).
- D. Bhat, S. Hauf, C. Plessy, Y. Yokobayashi, S. Pigolotti, "Speed variations of bacterial replisomes", Elife 11:e75884 (2022).
- H. Tsai, D.W. Carlson, A. Koldaeva, S. Pigolotti, A.Q. Shen, "Optimization and Fabrication of Multi-Level Microchannels for Long-Term Imaging of Bacterial Growth and Expansion", Micromachines 13(4), 576 (2022).
4.2 Books and other one-time publications
Nothing to report
4.3 Oral and Poster Presentations
- A.Koldaeva, I. Dubanevics. “Population genetics of E. coli in microchannels: theory, experiment and numerical simulation.”, Poster, Modeling on Ecology and Evolution Workshop (MEEW), October (2022)
- S. Pigolotti. "Speed variations of bacterial replisomes" presentation, The 3rd Workshop on Stochasticity and Fluctuations in Small Systems, Pohang Korea, November 22-25 (2022)
- F. Pflug. "Studying Bacterial Replication through Marker Frequency Analysis" presentation, The 3rd Workshop on Stochasticity ann Fluctuations in Small Systems, Pohang Korea, November 22-25 (2022)
- S. Pigolotti. "Speed variations of bacterial replisomes" Presenattion, APS March Meeting 2023, Las Vegas, USA. March 5-10 (2023).
- S. Pigolotti. "Dynamics of growing bacterial cells" Presetation, The 28th EAJS -East Asia Joint Symposium, OIST, Japan. October 26-28 (2022)
- S. Pigolotti. "How physical forces shape microbial biodiversity" Presentation, Tohoku University -OIST 3rd Joint Workshop on Biodiversity, Online, Japan, October 24-26 (2022)
- F. Pflug. "Neutral competition shapes the clonal composition of cerebral organoids", ISCO 2023 (Interdisciplinary Science Conference in Okinawa - Physics and Mathematics meet Medical Science), Okinawa, February 27-March 3 (2023)
- F. Pflug. "Studying Bacterial Replication Marker Frequency Analysis", University of Vienna, September (2022)
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Workshop "Cells, energetics, and information: New perspectives on nonequilibrium systems"
- Date: June 6-10, 2022
- Venue: Online
- Invited Speakers:
- Deepak Bhat (OIST)
- Massimiliano Esposito (U. Luxembourg)
- Nikta Fakhri (MIT)
- Chikara Furusawa (U. Tokyo, RIKEN)
- Jordan Horowitz (U. Michigan)
- Kyogo Kawaguchi (RIKEN)
- Akihiro Kusumi (OIST)
- Namiko Mitarai (U. Copenhagen)
- Arvind Murugan (U. Chicago)
- Armita Nourmohammad (U. Washington)
- Felix Ritort (U. Barcelona)
- Jonathan Rodenfels (MPI-CBG)
- Takahiro Sagawa (U. Tokyo)
- Satoshi Sawai (U. Tokyo)
- Pablo Sartori (Gulbenkian Institute)
- Amy Shen (OIST)
- David Sivak (U. Simon Fraser)
- Evelyn Tang (U. Rice)
- Tsvi Tlusty (IBS, Ulsan)
6.2 RNA-based ligation reactions driven by a microscale water cycle under CO2 atmosphere.
- Date: September 9, 2022
- Venue: OIST Campus Lab 4 E26
- Speaker: Leonie Karr
6.3 GuaCAMOLE: fragment GC bias-aware abundance estimation from metagenomic data increases accuracy and replicability
- Date: November 3, 2022
- Venue: OIST Campus Lab3 B712
- Speaker: Laurenz Holcik (Center of Integrative Bioinformatics, University of Vienna)
6.4 Termination of DNA replication in bacteria
- Date: December 7, 2022
- Venue: OIST Campus Lab 4 E26
- Speaker: Christian Rudolph
7. Other
Nothing to report.



