FY2014 Annual Report
G0 Cell Unit
Professor Mitsuhiro Yanagida

Abstract
The G0 Cell Unit studies how cells in the proliferative (dividing) or quiescent (non-dividing) phase respond to nutritional shifts (e.g., nitrogen source starvation and glucose limitation). We identify gene products (mostly proteins) and chemical factors (small molecules, metabolites) that affect adaptation mechanisms in order to understand the basis for longevity of long-term quiescent cells. We employ fission yeast as a eukaryotic cell model as the great majority of this organism’s genes are conserved in human. Our current principal research projects are:
(1a) Methodological development for metabolomic analysis.
(1b) Comprehensive and quantitative human blood metabolomics.
(2) Understanding cell regulation in response to nitrogen deprivation.
(3) Understanding cell regulation in response to glucose starvation.
(4) Understanding chromosomal regulatory mechanisms involving condensin and other nutrient adaptation-related nuclear proteins.
In FY2014, we published seven original and two review (or book chapter) articles on metabolomic comparison between human blood and fission yeast, ergothioneine biosynthetic pathway in fission yeast, major fission yeast glucose transporters, the role of condensin in proliferating and quiescent cells, and other topics, as introduced in 3. Activities and Findings and listed in 4. Publications.
1. Staff
- Dr. Takeshi Hayashi, Group leader
- Dr. Kazuki Kumada, Group leader
- Dr. Romanas Chaleckis, Researcher (from October)
- Dr. Xiaodong He, Researcher (until June)
- Dr. Norihiko Nakazawa, Researcher
- Dr. Tomas Pluskal, Researcher
- Dr. Kenichi Sajiki, Researcher
- Dr. Takayuki Teruya, Researcher
- Dr. Emily Tsang, Researcher
- Ms. Orie Arakawa, Technical staff
- Mr. Masahiro Ebe, Technical staff
- Ms. Ayaka Mori, Technical staff
- Ms. Yuria Tahara, Technical staff
- Ms. Junko Takada, Technical staff (from July)
- Ms. Risa Uehara, Technical staff
- Ms. Li Wang, Technical staff
- Ms. Chikako Sugiyama, Research administrator
- Ms. Caroline Starzynski, OIST Student
- Mr. Xingya Xu, Special Research Student
- Ms. Joanne Becker, Research Intern (from June, until August)
- Ms. Hannah Furgason, Research Intern (from June, until August)
- Ms. Zygimante Tarnauskaite, Research Intern (from July, until September)
- Mr. Zenan Wang, Research Intern (from January)
2. Collaborations
- Theme: Metabolite structure determination using NMR
- Type of collaboration: Joint research
- Researchers:
- Dr. Masaru Ueno, Department of Molecular Biotechnology, Graduate school of Advanced Science of Matter, Hiroshima University
- Theme: Screening for low-glucose sensitive mutants in fission yeast
- Type of collaboration: Joint research
- Researchers:
- Professor Shigeaki Saitoh, Institute of Life Science, Kurume University
- Theme: Identification of the molecular mechanism, required for the maintenance of cell viability in low glucose condition
- Type of collaboration: Joint research
- Researchers:
- Professor Kunihiro Ohta, Department of Life Science, Graduate School of Arts and Science, The University of Tokyo
- Theme: Analysis of biomarkers, discovered by research using S.pombe, in human blood
- Type of collaboration: Joint research
- Researchers:
- Dr. Hiroshi Kondoh, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Dr. Takumi Mikawa, Department of Geriatric Medicine, Graduate School of Medicine, Kyoto University
- Theme: Functional analysis of condensin complex in the regulation of chromosome segregation and gene expression
- Type of collaboration: Joint research
- Researchers:
- Dr. Takashi Sutani, Institute of Molecular Cellular Biosciences, The University of Tokyo
- Theme: Physiological/genetic analysis of cellular adaptation strategy to environmental changes
- Type of collaboration: Joint research
- Researchers:
- Dr. Kojiro Takeda, Department of Biology, Faculty of Science and Engineering, Konan University
- Theme: Metabolic analysis of active ingredients of health foods
- Type of collaboration: Joint research
- Researchers:
- Professor Hiroaki Masuzaki, Division of Endocrinology, Diabetes and Metabolism, Hematology, Rheumatology (Second Department of Internal Medicine), Graduate School of Medicine, University of the Ryukyus
3. Activities and Findings
3.1 Unexpected similarities between the Schizosaccharomyces and human blood metabolomes, and novel human metabolites
Metabolomics, a modern branch of chemical biology, provides qualitative and quantitative information about the metabolic states of organisms or cells at the molecular level. Numerous blood metabolomic analyses have been previously reported, mostly of serum and plasma. However, metabolomics of whole blood or red blood cells (RBCs) have been less well investigated, except for several reports on long-term stored blood, blood of disease patients, or blood marker compounds of food intake. This study focuses on the metabolome of human blood cells, mainly RBCs, separated from plasma by low speed centrifugation. We performed LC-MS-based metabolomic analysis of human blood, plasma, and RBCs in comparison with previously published metabolomic results from the fission yeast, Schizosaccharomyces pombe. The two metabolomic datasets were highly similar: 101 of 133 compounds identified in human blood (75%) were also present in S. pombe, and 45 of 57 compounds enriched in red blood cells (RBCs) (78%), were also present in yeast (Figure 1). The most abundant metabolites were ATP, glutathione, and glutamine. Apart from these three, the next most abundant metabolites were also involved in energy metabolism, anti-oxidation, and amino acid metabolism. The high similarity of these two metabolomes raises the possibility that S. pombe genetics might be useful to understand the role of certain metabolites, such as small anti-oxidants (ophthalmic acid, ergothioneine, and glutathione), which are enzymatically synthesized in S. pombe.
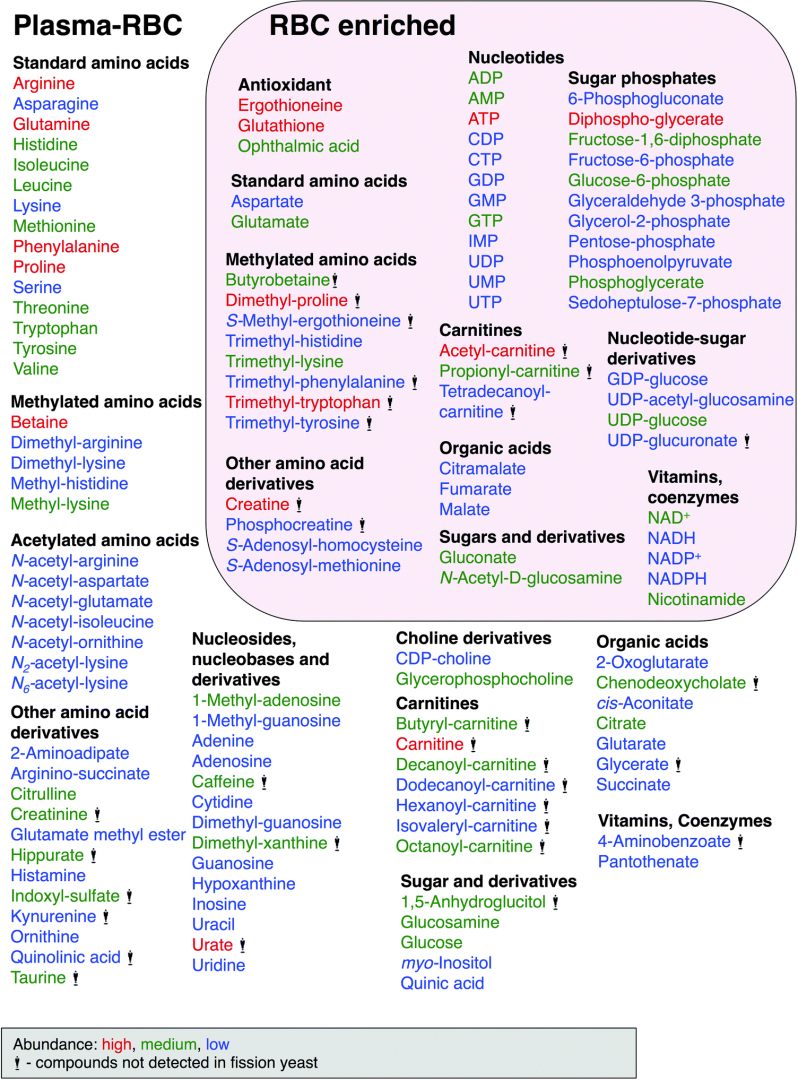
Figure 1: Human blood compounds identified and characterized in this study. Abundance of compounds classified by peak area size, indicated by color, red (high), green (medium) and blue (low). Compounds with the statue symbol are not present in S. pombe.
To our knowledge, 14 metabolites have not hitherto been reported in human blood, based on a recent report of detected blood metabolites and literature database searches. These new blood metabolites include citramalate, dimethyl-lysine, GDP-glucose, glutamate methyl ester, N-acetyl-glutamate, N-acetyl-(iso)leucine, N2-acetyl-lysine, N6-acetyl-lysine, trimethyl-histidine, trimethyl-phenylalanine, trimethyl-tryptophan, trimethyl-tyrosine, UDP-acetyl-glucosamine and UDP-glucuronate. The eight compounds in boldface were enriched in RBCs, while the ten compounds in italics were also found in S. pombe. Ten of the 14 novel blood metabolites are methylated or acetylated amino acids. This study was performed in collaboration with Dr. Hiroshi Kondoh (Kyoto University) and published in Molecular BioSystems (Chaleckis et al., 2014).
3.2 Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in fission yeast
Ergothioneine (EGT) is a small, sulfur-containing metabolite (229 Da) synthesized by various species of bacteria and fungi, which can accumulate to millimolar levels in tissues or cells (e.g. erythrocytes) of higher eukaryotes. It is commonly marketed as a dietary supplement due to its proposed protective and antioxidative functions. However, so far, no rigorous research has conclusively demonstrated any benefit of EGT in vivo. It is unclear whether EGT consumption contributes to human health, and if it does, what daily intake is optimal. In this study we report the genes forming the two-step EGT biosynthetic pathway in S. pombe (Figure 2). We identified the first gene, egt1+ (SPBC1604.01), by sequence homology to previously published genes from Neurospora crassa and Mycobacterium smegmatis. We showed, using metabolomic analysis, that the ∆egt1 deletion mutant completely lacked EGT and its precursors (trimethyl histidine/hercynine and hercynylcysteine sulfoxide). Since the second step of EGT biosynthesis has not been characterized in eukaryotes, we examined four putative homologs (Nfs1/ SPBC21D10.11c, SPAC11D3.10, SPCC777.03c, and SPBC660.12c) of the corresponding mycobacterial enzyme EgtE. Among deletion mutants of these genes, only one (∆SPBC660.12c, designated ∆egt2) showed a substantial decrease in EGT, accompanied by accumulation of its immediate precursor, hercynylcysteine sulfoxide.
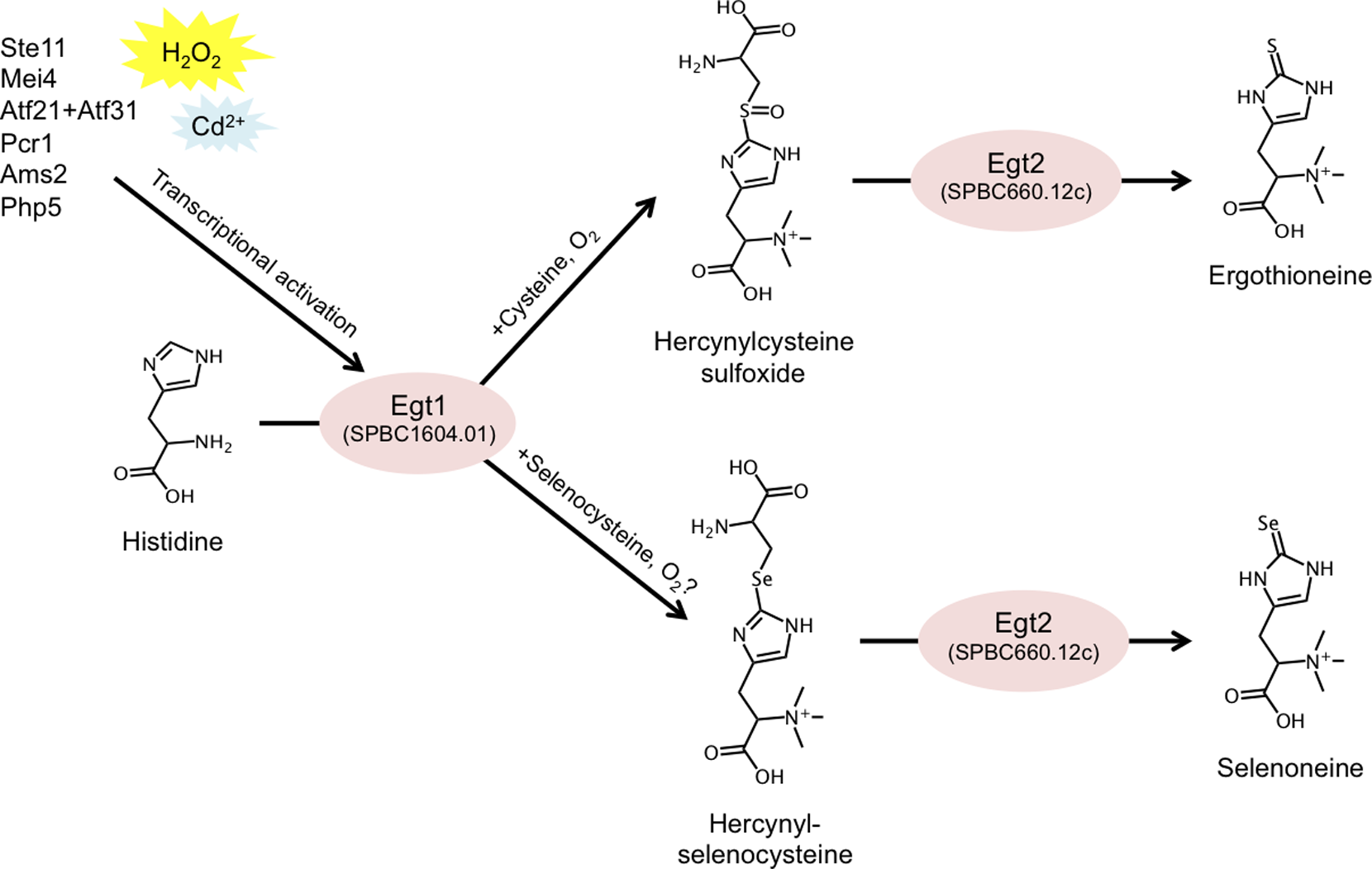
Figure 2: Summary of the described EGT and selenoneine biosynthetic pathway in S. pombe, and its transcriptional regulation.
EGT-deficient strains exhibited no phenotypic defects during vegetative growth or quiescence. To effectively study the role of EGT, we constructed an egt1+ overexpression system by replacing its native promoter with the nmt1+ promoter, which is inducible in the absence of thiamine. We employed three versions of the nmt1 promoter with increasing strength of expression and confirmed corresponding accumulations of EGT. We quantified the intracellular concentration of EGT in S. pombe (0.3, 157.4, 41.6, and up to 1606.3 µM in vegetative, nitrogen-starved, glucose-starved, and egt1+-overexpressing cells, respectively) and described its gradual accumulation under long-term quiescence. Finally, we demonstrated that the EGT pathway can also synthesize selenoneine, a selenium-containing derivative of EGT, when the culture medium is supplemented with selenium. We further found that selenoneine biosynthesis involves a novel intermediate compound, hercynylselenocysteine. We propose that genetic and metabolomic analyses, together with the collection of S. pombe strains introduced in this study, may provide ideal tools to further investigate the in vivo role of EGT. This study was published in PLoS ONE (Pluskal et al., 2014).
3.3 Mechanisms of expression and translocation of major fission yeast glucose transporters regulated by CaMKK/phosphatases, nuclear shuttling, and TOR
Hexose transporters are required for cellular glucose uptake; thus they play a pivotal role in glucose homeostasis in multicellular organisms. For proper levels of hexose uptake, expression levels and activity of each transporter must be modulated according to changes in environmental hexose concentrations and/or the physiologic requirement for hexose in tissues. An understanding of the mechanisms involved in controlling the expression and activity of the transporters is therefore important. In this study, we comprehensively analyzed eight hexose transporters encoded by ght1+–ght8+ genes in the fission yeast S. pombe. Despite the high similarity in their sequences, the eight transporters do not appear to function redundantly. Among them, ght5+ is essential and sufficient for cell proliferation under low-glucose conditions.
The high-affinity transporter Ght5 is regulated with regard to transcription and localization, much like the human GLUT transporters, which are implicated in diabetes. When restricted to a glucose concentration equivalent to that of human blood, the fission yeast transcriptional regulator Scr1, which represses Ght5 transcription in the presence of high glucose, is displaced from the nucleus. Its displacement is dependent on Ca2+/calmodulin-dependent kinase kinase, Ssp1, and Sds23 inhibition of PP2A/PP6-like protein phosphatases. Newly synthesized Ght5 locates preferentially at the cell tips with the aid of the target of rapamycin (TOR) complex 2 and Gad8/Akt signaling. These results provide a novel view that a series of large-scale changes, such as de novo synthesis of Ght5 and its preferential localization at the cell tip, are induced in S. pombe during cellular adaptation to low-glucose conditions (Figure 3). Our findings shed light on the molecular mechanism in fission yeast that modulates glucose uptake and consumption, depending on the glucose concentration in the cellular environment, which is presumed to be crucial for maintaining glucose homeostasis in multicellular organisms. This study was performed in collaboration with Professor Shigeaki Saitoh (Kurume University) and published in Molecular Biology of the Cell (Saitoh et al., 2015).
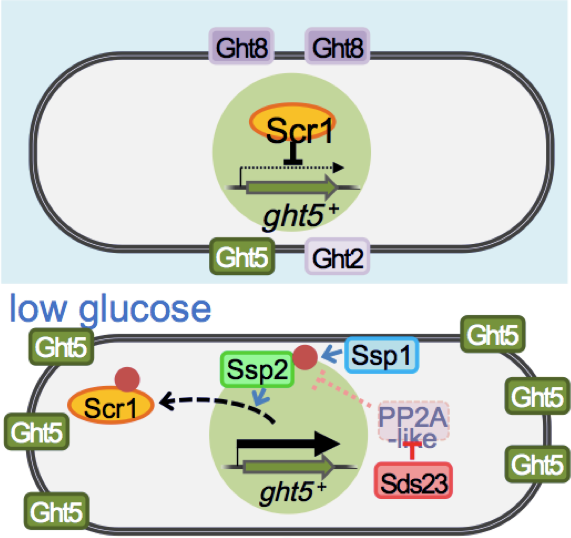
Figure 3: Transcription of the major glucose transporter (Ght5) is up-regulated by CaMKK-like Ssp1, and PP2A-like phosphatase inhibitor Sds23 through the nuclear exclusion of repressor Scr1 under low-glucose conditions. On the other hand, the accumulation of newly synthesized Ght5 at the cell tips requires TORC2-AKT signaling.
3.4 ATPase-dependent auto-phosphorylation of the open condensin hinge diminishes DNA binding
Condensin, which contains two structural maintenance of chromosome (SMC) subunits and three regulatory non-SMC subunits, is essential for many chromosomal functions, including mitotic chromosome condensation and segregation. The ATPase domain of the SMC subunit comprises two termini connected by a long helical domain that is interrupted by a central hinge. The role of the ATPase domain has remained elusive. In this study, we employed an in vitro approach, building upon our previous study, which demonstrated the ability of the condensin SMC heterodimer Cut3– Cut14 to remove single-stranded (ss) DNA-binding protein RPA or Ssb1, which had been bound to the unwound ssDNAs. As the elimination of protein and/or RNA during the re-annealing reaction per se did not require ATP, we wondered how ATP interacts with condensin’s ATPase domain during the re-annealing reaction. To our great surprise, we discovered that the phosphate groups of ATP bind to multiple sites on Cut3, an SMC4-like subunit, but this apparent ‘auto-phosphorylation’ is greatly diminished when the holocondensin complex, which has ample ATPase activity, is employed. We show that multiple phosphorylation sites are located in the conserved, glycine-rich stretch at the hinge interface surrounded by the highly basic DNA-binding patch, and that phosphorylation is abolished in ATPase mutants. Phosphorylation reduces affinity for DNA. Consistently, phosphomimetic mutants produce severe mitotic phenotypes.
Structural evidence suggests that prior opening (though slight) of the hinge is necessary for phosphorylation, which is implicated in condensin’s dissociation from and its progression along DNA. We propose that hinge phosphorylation represents a step in condensin’s ATPase cycle, and that it is important for understanding condensin’s dissociation from chromosomal DNA (Figure 4). In other words, ATP enables the mobility of condensin along chromosomes by causing it to dissociate from DNA. This study was performed in collaboration with Professor Chikashi Toyoshima (The University of Tokyo) and published in Open Biology (Akai et al., 2014).
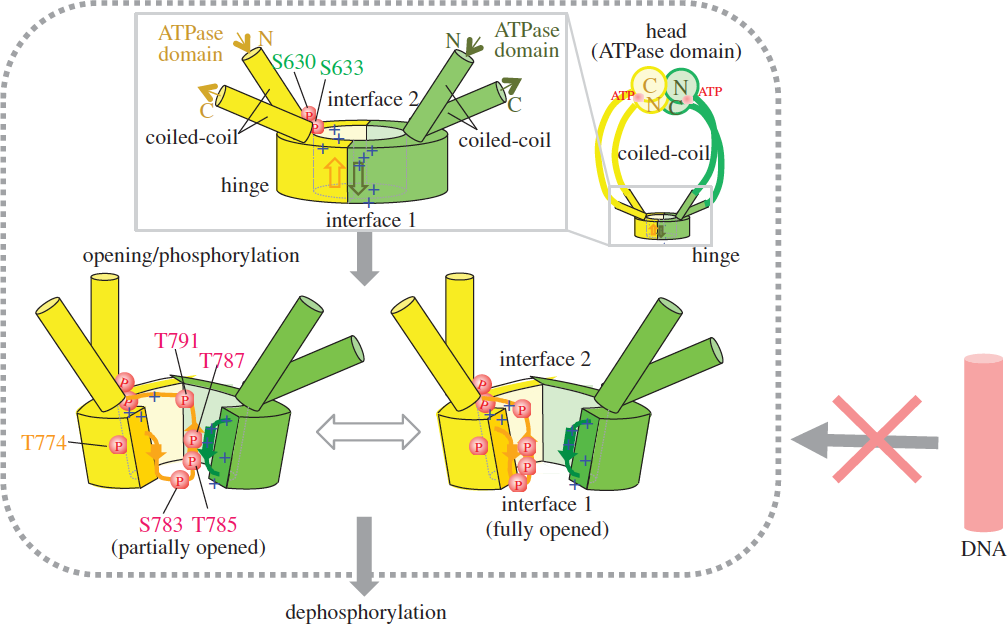
Figure 4: Schematized opening, (auto)phosphorylation and dephosphorylation of the hinge. Schematized whole condensin shown at right top. The phosphorylated hinge is presumed to be unable to bind ssDNA (red DNA rod with the crossed arrow). This hypothesis is based on two crystal structures (closed and opened) of the hinge. Hinges of Cut3/SMC4 (in yellow) and Cut14/SMC2 (in green) are illustrated, showing locations of thiophosphorylated residues.
3.5 Condensin HEAT subunits required for DNA repair, kinetochore/centromere function and ploidy maintenance in fission yeast
Condensin, a central player in eukaryotic chromosomal dynamics, contains five evolutionarily-conserved subunits. Two SMC (structural maintenance of chromosomes) subunits contain ATPase, hinge, and coiled-coil domains. One non-SMC subunit is similar to bacterial kleisin, and two other non-SMC subunits contain HEAT (similar to armadillo) repeats. Genetic dissections of condensin’s non-SMC subunit functions have been scarce. To characterize these subunits in a systematic way, we attempted to construct mutants for all 3 subunits using error-prone PCR mutagenesis. We were able to obtain and characterize 21 newly isolated mutants of cnd1, cnd2, and cnd3, containing only single amino acid substitutions. The novelty of our approach for isolating condensin non-SMC mutants was that we did not screen the microscopic condensation-defect, which was used as the selective phenotype in previous studies. In this study we isolated mutants that produced ts and/or DNA damage-sensitive phenotypes.
Beside previously known mitotic functions in chromosome condensation and segregation and in DNA repair, we identified two novel condensin functions. First, HEAT repeat-containing subunits, Cnd1 and Cnd3, are involved in maintaining the haploid genome state. Each of the three cnd1 and the three cnd3 mutations resided in or near the HEAT repeats caused an increase in cell size and doubled DNA content (Figure 5). These mutants normally proliferate, but do not form viable spores in meiotic segregation when crossed with wild-type haploids, probably due to triploid meiosis producing massive aneuploidy. Condensin thus seems to be required for maintaining the ploidy level via its HEAT-containing subunits. Secondly, we show that one cnd3-L269P mutant is defective in centromere/kinetochore function and spindle assembly checkpoint control. Consistently, this mutant, bearing an altered residue in the conserved HEAT repeat is hypersensitive to TBZ, a tubulin poison. These results together demonstrate novel aspects of the role of non-SMC, HEAT-containing subunits of condensin. The non-SMC condensin subunits Cnd1 and Cnd3 may play an unexpected role in cell growth and/or ploidy level stability. This study was published in PLOS One (Xu et al., 2015).
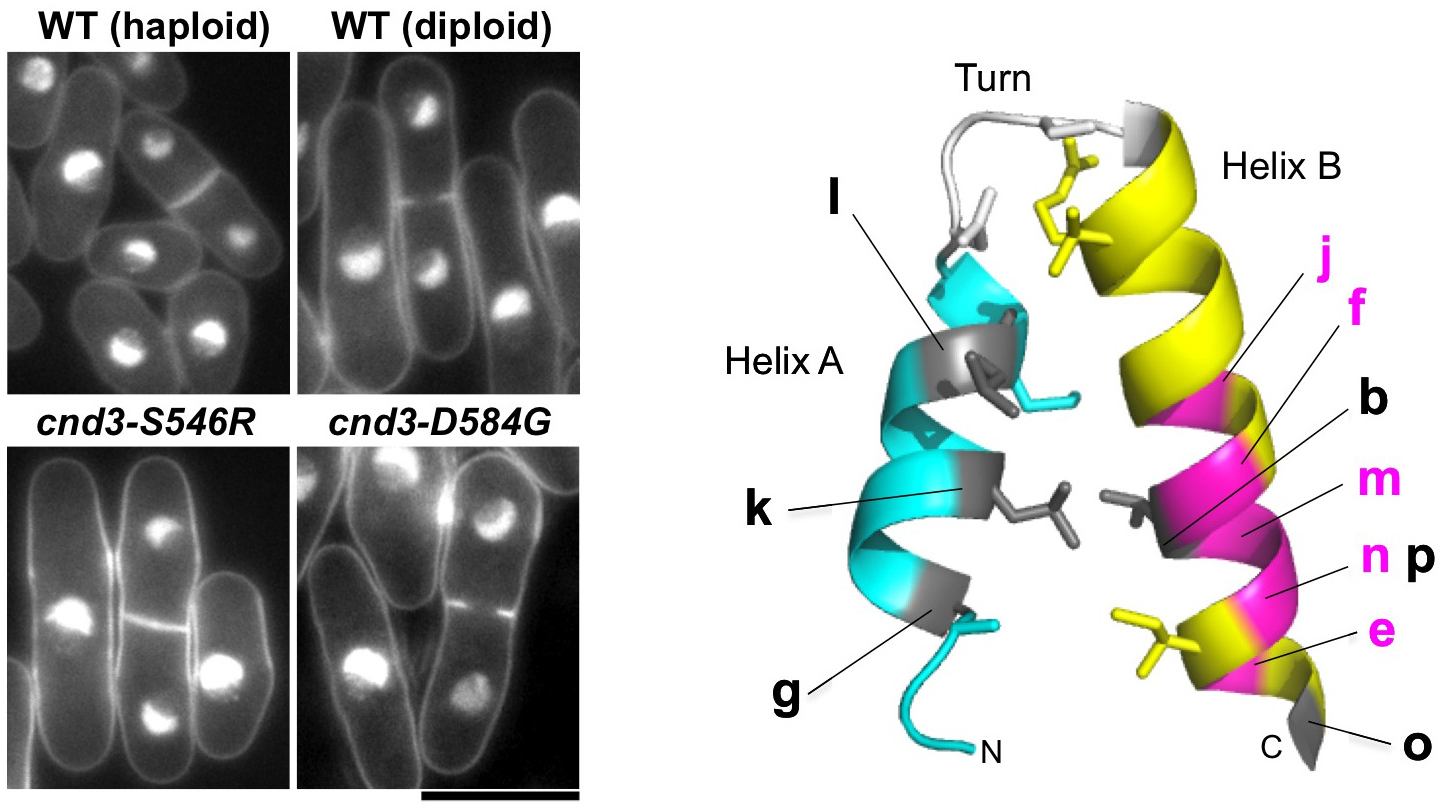
Figure 5: Condensin non-SMC subunits affect chromosome ploidy (left: mutants of non-SMC subunits, right: mutation sites of cnd1 and cnd3 on conserved HEAT-repeat structure).
3.6 Fission yeast centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex
CENP-A is a centromere-specific variant of histone H3 that is required for accurate chromosome segregation. The Mis18 complex is hypothesized to have a role in centromere priming for CENP-A deposition in the subsequent cell cycle, and thus, histone acetylation is suggested to be involved in Mis18 complex function. The molecular function of the Mis18 complex, however, remains unclear. In this study, we investigated the molecular organization of the Mis16-Mis18 complex in fission yeast and identified two novel Mis18-interacting proteins, Mis19 and Mis20. Our findings from two- and three-hybrid analyses as well as mass spectrometry support the notion that Mis16, Mis18, Mis19 and Mis20 form a complex and Mis16 forms another complex with Hat1 (Figure 6A). Our analyses also indicated that Mis19 acts as a bridge between Mis16 and Mis18 in the Mis18 complex.
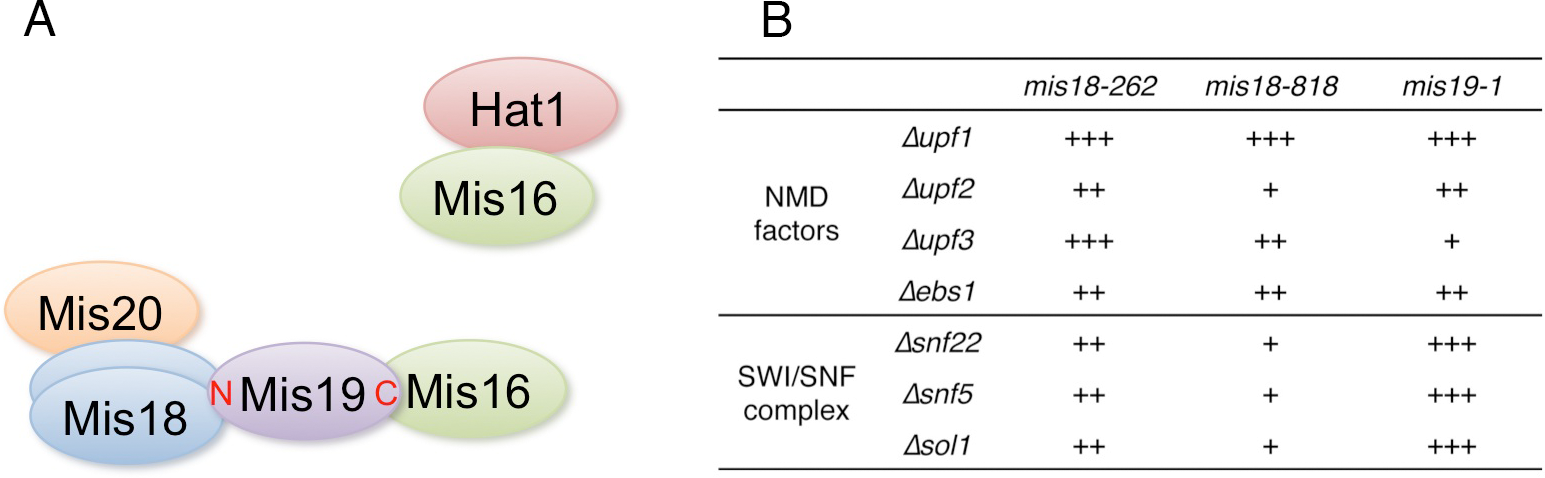
Figure 6: A. Summary of the physical interactions of Mis16- and Mis18-interacting proteins. B. Summary of suppression of mis18 and mis19 mutants by deletion mutants of NMD factors and SWI/SNF complex components. +++ is strongest suppression, and + is weak suppression.
In addition, we screened extragenic suppressors of mis18 and mis19 mutants and identified the functional link between the Mis18 complex and non-sense-mediated mRNA decay (NMD) factors or SWI/SNF chromatin-remodeling complex (Figure 6B). NMD is an evolutionarily conserved RNA surveillance system that degrades mRNAs with premature termination (non-sense) codons and also suggested to participate in the general regulation of gene expression, affecting the expression of genes lacking premature termination codons. SWI/SNF chromatin-remodeling complex has a critical role in transcription. Our results suggest that the Mis16-Mis18-Mis19-Mis20 CENP-A-recruiting complex, which is functional in the G1-S phase, may be counteracted by the SWI/SNF chromatin-remodeling complex and NMD, which may prevent CENP-A deposition at the centromere. In short, our findings not only revealed the organization of the Mis18 complex in fission yeast, but also provide some insight into the function of the Mis18 complex, which might be involved in centromeric transcription or noncoding RNAs from the centromere core domain. This study was published in Genes to Cells (Hayashi et al., 2014).
4. Publications
4.1 Journals
- Hayashi, T., Ebe, M., Nagao, K., Kokubu, A., Sajiki, K. & Yanagida, M. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes to cells, doi:10.1111/gtc.12152 (2014).
- Saitoh, S. & Yanagida, M. Does a shift to limited glucose activate checkpoint control in fission yeast? FEBS letters, doi:10.1016/j.febslet.2014.04.047 (2014).
- Pluskal, T., Ueno, M. & Yanagida, M. Genetic and Metabolomic Dissection of the Ergothioneine and Selenoneine Biosynthetic Pathway in the Fission Yeast, S. pombe, and Construction of an Overproduction System. PLoS One 9, e97774, doi:10.1371/journal.pone.0097774 (2014).
- Chaleckis, R., Ebe, M., Pluskal, T., Murakami, T., Kondoh, H. & Yanagida, M. Unexpected similarities between the Schizosaccharomyces and human blood metabolomes, and novel human metabolites. Mol Biosyst, doi:10.1039/C4MB00346B (2014).
- Saitoh, S., Mori, A., Uehara, L., Masuda, F., Soejima, S. & Yanagida, M. Mechanisms of expression and translocation of major fission yeast glucose transporters regulated by CaMKK/phosphatases, nuclear shuttling and TOR. Molecular biology of the cell, doi:10.1091/mbc.E14-11-1503 (2015).
- Murakami, I., Chaleckis, R., Pluskal, T., Ito, K., Hori, K., Ebe, M., Yanagida, M. & Kondoh, H. Metabolism of Skin-Absorbed Resveratrol into Its Glucuronized Form in Mouse Skin. PLoS One 9, e115359, doi:10.1371/journal.pone.0115359 (2014).
- Akai, Y., Kanai, R., Nakazawa, N., Ebe, M., Toyoshima, C. & Yanagida, M. ATPase-dependent auto-phosphorylation of the open condensin hinge diminishes DNA binding. Open Biology, doi:10.1098/rsob.140193 (2014).
- Xu, X., Nakazawa, N. & Yanagida, M. Condensin HEAT Subunits Required for DNA Repair, Kinetochore/Centromere Function and Ploidy Maintenance in Fission Yeast. PLoS One 10, e0119347, doi:10.1371/journal.pone.0119347 (2015).
4.2 Books and other one-time publications
- Yanagida, M. Mitosis The Role of Model Organisms in the History of Mitosis Research, in Cold Spring Harbor Perspectives in biology, Mitosis, Cold Spring Harbor Laboratory Press (2014).
- Pluskal, T. Comprehensive metabolomic analysis of the fission yeast, Schizosaccharomyces pombe Doctor of Science thesis (2014).
4.3 Oral and Poster Presentations
- Yanagida, M. Human blood metabolome profiling: Identification of individually-variable and age-implicated compounds, in 50th Year Anniversary Symposium Molecular Biology Dept. of the University of Geneva (2014).
- Yanagida, M. Protein dephosphorylation in regulating glucose metabolism in 11th International Conference on Protein Phosphatase, Tohoku University, Sendai (2014).
- Yanagida, M. A new era for condensin study: transcription and structural biology, in Institute of Molecular Biology Symposium, Institute of Molecular Biology, Taipei, Taiwan (2014).
- Yanagida, M. Does low glucose adaptation checkpoint exist?, in FEBS/EMBO2014, Paris, France (2014).
- Pluskal, T. Genetic and metabolomic dissection of the ergothioneine biosynthetic pathway in the fission yeast, S. pombe, in Metabolomics 2014, Tsuruoka, Yamagata (2014).
- Pluskal, T. Combined use of genetics and metabolomics to study the biosynthesis and function of small antioxidative metabolites in S. pombe, in FEBS/EMBO 2014, Paris, France (2014).
- Sajiki, K. Metabolomic Analysis of Fission Yeast at the Onset of Nitrogen Starvation, in FEBS/EMBO2014, Paris, France (2014).
- Nakazawa, N., Xu, X., Sajiki, K., Arakawa, O. & Yanagida, M. Condensin relieves the obstructive effect of transcription on chromosome segregation in fission yeast, in The 27th Yeast Genetic Forum, University of Tokyo, Japan (2014).
- Pluskal, T. Compounds implicated in cellular quiescence revealed by fission yeast metabolomics, Cambridge University, United Kingdom (2014).
- Pluskal, T. Compounds implicated in cellular quiescence revealed by fission yeast metabolomics, Dana-Farber Cancer Institute / Harvard Medical School, Boston, USA (2015).
- Pluskal, T. Compounds implicated in cellular quiescence revealed by fission yeast metabolomics, Whitehead Institute for Biomedical Research, Cambridge, USA (2015).
- Pluskal, T. Compounds implicated in cellular quiescence revealed by fission yeast metabolomics, Harvard T.H. Chan School of Public Health, Boston, USA (2015).
- Yanagida, M. Large scale cell re-modeling at nucleo-cytoplasm-cell tip in fission yeast ensures division in low-glucose, University of Fribourg, Switzerland (2014).
- Yanagida, M. Large scale cell re-modeling ensures division under starvation in fission yeast, Cancer Research UK, London (2014).
- Yanagida, M. Condensin versus Transcription and Processing Machinery, University of Cambridge, UK (2014).
- Yanagida, M. Mechanisms of cell remodeling in response to nutritional starvation, Graduate School of Frontier Biosciences, Osaka University (2015).
5. Intellectual Property Rights and Other Specific Achievements
Nothing to report
6. Meetings and Events
6.1 Seminar
- Title: Mechanisms how Parkinson’s disease-related glyoxalase DJ-1 supports mitochondrial membrane potential and in vitro survival of dopaminergic neurons
- Date: April 22, 2014
- Venue: C210, OIST Campus Center Building
- Speaker: Dr. Yusuke Toyoda (Max Planck Institute of Molecular Cell Biology and Genetics)
6.2 Seminar
- Title: Searching for cancer’s new weak points using fission yeast
- Date: January 21, 2015
- Venue: C016, OIST Campus Lab1
- Speaker: Dr. Masaru Ueno (Graduate School of Advanced Sciences of Matter, Hiroshima University)
7. Other
Nothing to report.



