Garth Ilsley
Science and Technology Associate
1. Introduction
The focus of the past year has been on measuring and validating gene expression at the single cell level in Ciona intestinalis, a marine chordate, along with developing and testing models of enhancer regulation in Drosophila melanogaster.
2. Activities and Findings
Single-cell measurements in Ciona intestinalis
It is important to have measurements of transcription factor levels and gene outputs at single-cell resolution. Spatially averaging transcription factor measurements obscures cellular state and hence limits the effectiveness of statistical studies of the regulatory inputs and outputs in the cell. Therefore, to understand gene regulation it is crucial to develop approaches for measuring gene expression at the single cell level.
In collaboration with the Luscombe Unit, I have been analysing RNA-seq data that we have generated from single cells in Ciona intestinalis. These data have allowed us to quantify expression at single cell resolution in the early embryo. Our initial analysis indicates that our results are consistent with previous in situ hybridisation data, and will thus allow us to study gene expression comprehensively in the early embryo.
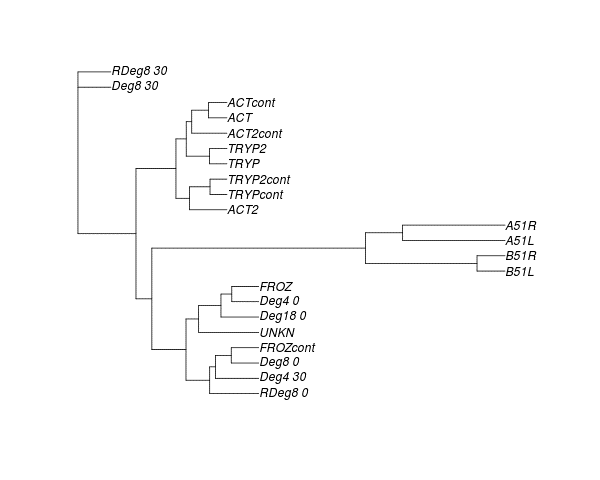
Figure 1: Our expression measurements cluster according to cell type and preparation protocol
Modelling transcriptional enhancer function in Drosophila
In collaboration with Justin Crocker and David Stern, we have examined the regulatory function of even-skipped enhancers in Drosophila. We have made use of modern imaging techniques to measure gene expression at a high level of resolution, thus enabling us to compare the results of model predictions of enhancer function with experimental perturbations, particularly those using transcription activator–like effectors fused to activators or repressors (TALEAs and TALERs).
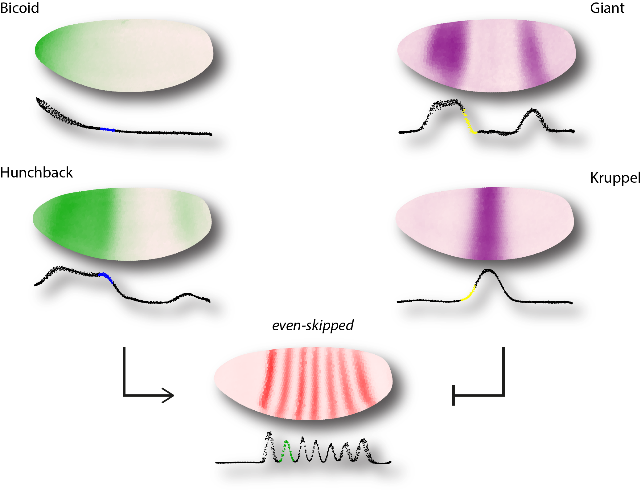
Figure 2: Data from the Berkeley Drosophila Transcription Network Project were used to model even-skipped regulatory function
3. Collaborations
Theme: Measuring gene expression with single-cell RNA-seq
Type of collaboration: Joint Research
Researchers: Luscombe Unit (OIST)
Theme: Modelling transcriptional enhancer function in Drosophila
Type of collaboration: Joint Research
Researchers: Justin Crocker and David Stern (Janelia Research Campus, USA)
4. Publications and other output
Garth R. Ilsley, Jasmin Fisher, Rolf Apweiler, Angela H. DePace, Nicholas M. Luscombe. A simple quantitative model of gradient interpretation can explain the precise pattern of even skipped in the Drosophila embryo. Cell Press Symposia: Transcriptional Regulation in Development, Chicago, USA. (July 2014).





